Team:Freiburg Bioware/NoteBook/Labjournal/September
From 2010.igem.org
(→123. labday 18.09.2010) |
(→Test digestion of pSB1C3_ZEGFR:1907_VP2/3) |
||
| Line 5,642: | Line 5,642: | ||
*<b>Expected fragment sizes:</b> <br/> | *<b>Expected fragment sizes:</b> <br/> | ||
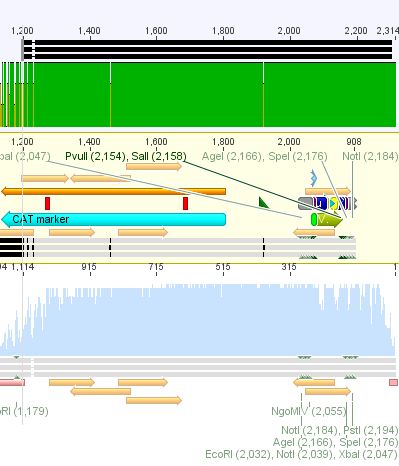
| - | + | P516/517: 3540bp / 651bp<br/> | |
| - | + | P514/515: 1300bp / 2700bp<br> | |
| + | P518-521: 2000bp / 700bp <br> | ||
| + | |||
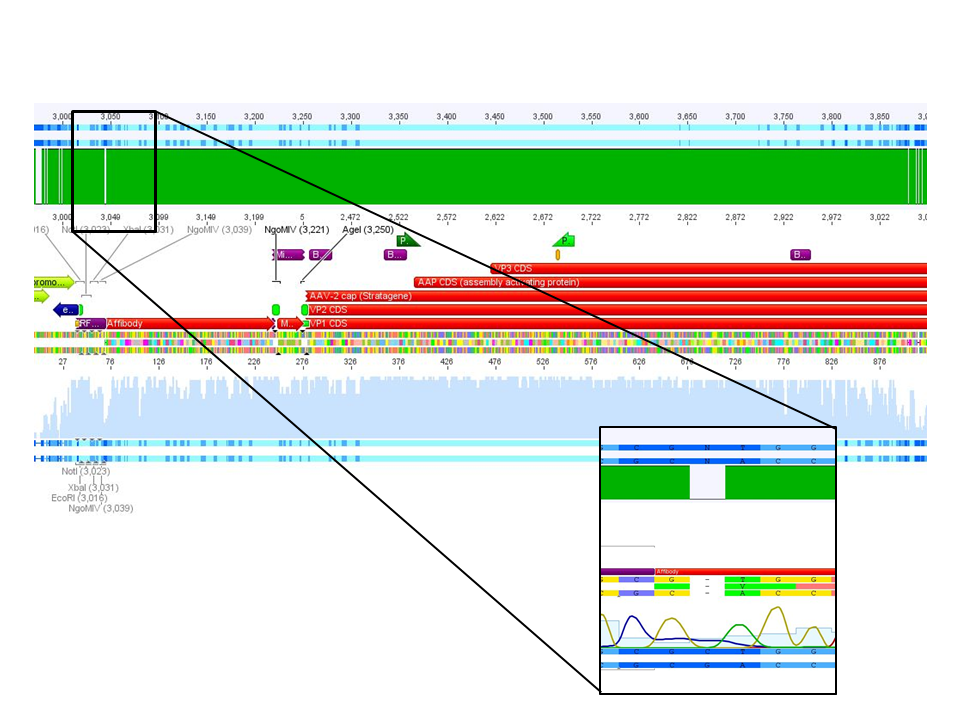
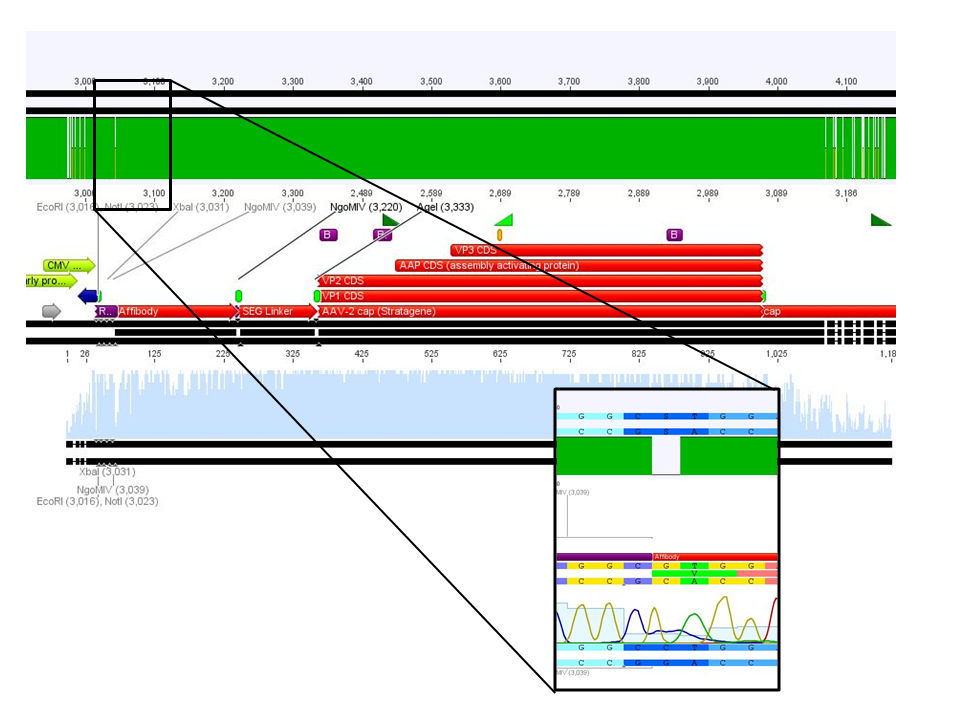
<p style="font-size:13px; color:#003399;"><b>Comment</b>: Sample was sent for sequencing with primer . </p> | <p style="font-size:13px; color:#003399;"><b>Comment</b>: Sample was sent for sequencing with primer . </p> | ||
Revision as of 12:48, 18 September 2010

September
107. labday 01.09.2010
Loop insertion BioBricks: Sequencing results
Investigator: Achim, Anna
Comment: Sequencing results of Loop insertion BioBricks from 31.08.10; the following clones were sent for sequencing:
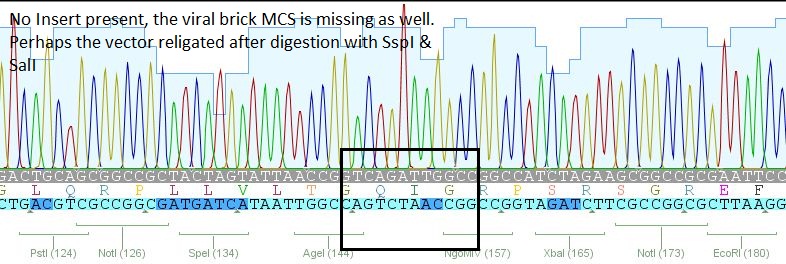
- 2: 453 BAP: No Insert
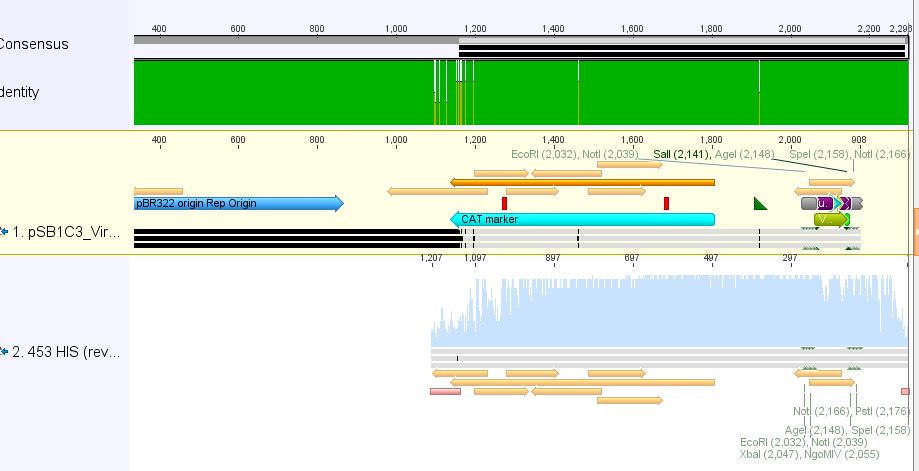
- 3:453 His: Insert okay
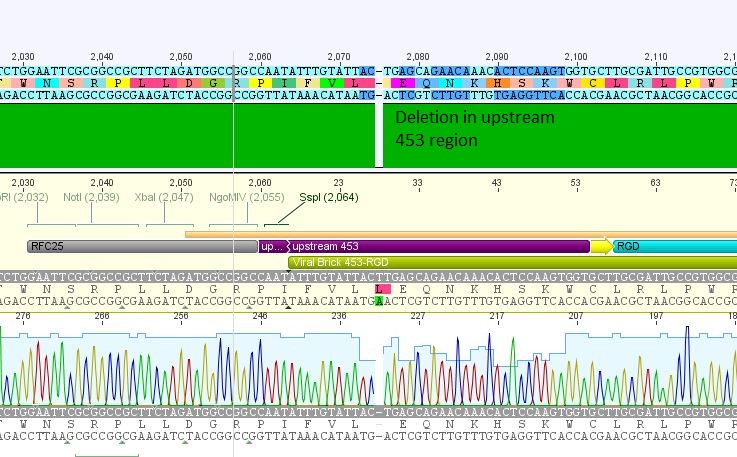
- 5: 453 RGD: Mutation in 453 upstream region
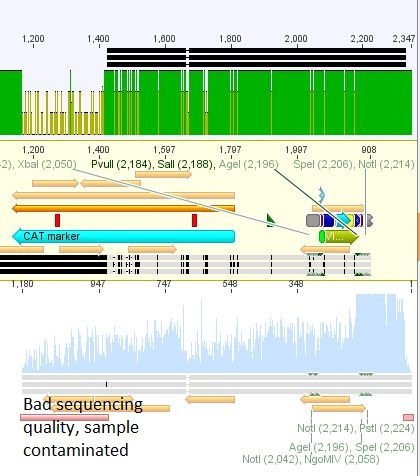
- 7: 587 BAP: Mutation in Bap insert
- 9:587 His: Insert okay
- 11:587 KO BAP : Insert okay
- 13:587 KO empty : Insert okay
- 16: 587 KO his: Mutation in 587 KO
- 17: 587 KO rgd: No insert
- 19 :587 RGD: Insert okay
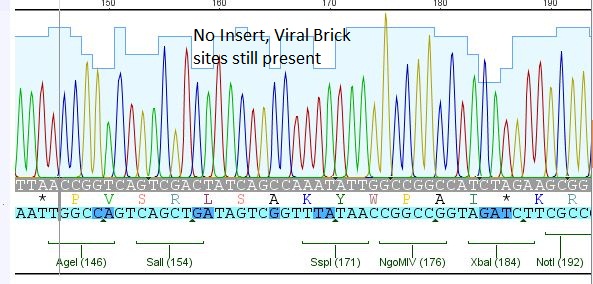
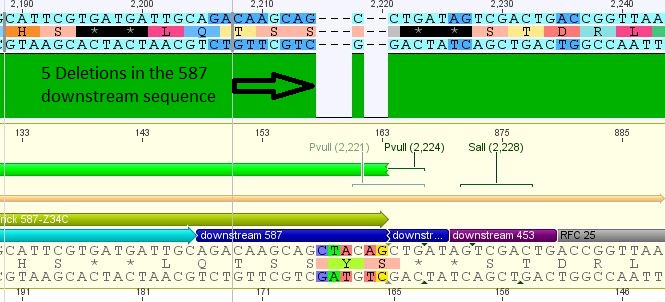
- 25: 587 Z34C: Mutation in downstream 587 region
Mini-Preps:
New Mini-Preps of the samples with no/mutated insert were prepared. The following clones were used:
| Components | 453 BAP | 587 BAP | 453 RGD | 587 KO RGD | 587 KO HIS | 453 Z34C | 587 Z34C | 587 KO Z34C | 587 KO Z34C SPACER |
| Clone | 1.3 + 1.4 | 2.3 + 2.4 | 4.3 + 4.4 | 6.3 + 6.4 | 9.3 + 9.4 | 11.3 + 11.4 | 12.2 + 12.4 | 13.2 + 13.4 | 14.3 + 14.4 |
Comment: Samples 1.3, 2.3, 4.3, 6.3, 9.3, 11.3, 12.2, 13.2, 14.3 were sent for sequencing. For sequencing results go to labday 02.09.10.
. Design of primers for pAAV_RC
Investigator: Bea
Primers for different cloning strategies of the synthesized cap gene into pAAV_Rc have been designed.
- The first primers are mutagenesis primers which delete the KpnI recognition site at position 3968. These primers can (or should be used) be used if cloning with BspMI/BfuI (isochizomer) does not cut the pAAV_RC efficiently. Instead of using this enzyme we can use XcmI to subclone the cap gene with performing a site-directed mutagenesis afterwards with designed primers.
- The second idea is to design oligoduplexes (hybridized oligos) which contain the recognition site for BspMI. In Gormley et al (2002) the idea was to add oligodupexes with the recognition site for BspMI and therefore provide the "second recognition site" in trans.
Next step: Check primers and order them today! Designed primers are stored in my Workingfolder under paav_rc and a word document was created whcih can be found under Oligos --> primers for different strategies...
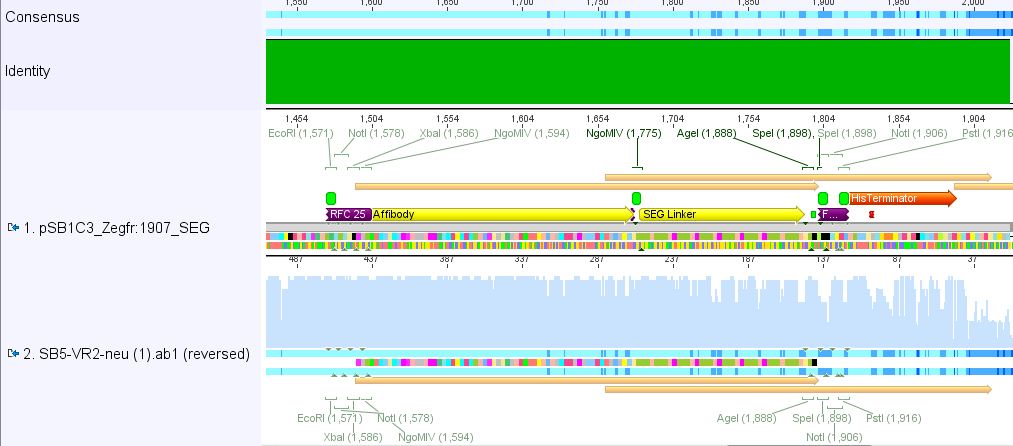
Sequencing results of pSB1C3_Zegfr:1907_Linker
Investigator: Hanna
Comment: Sequencing results looked all well. One bp was missing in the theoretical sequence of SEG suffix - but is correct in the actual sequence.
pSB1Cr_Zegfr:1907_MiddleLinker:
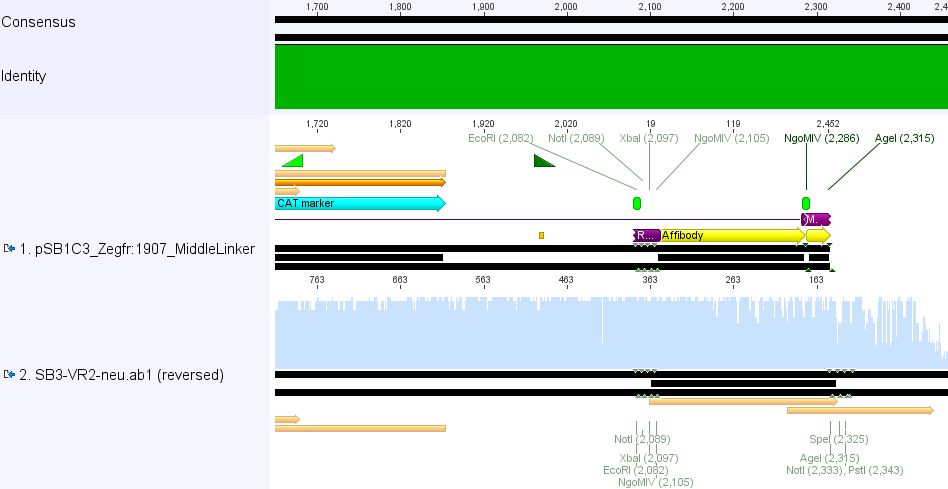
pSB1Cr_Zegfr:1907_ShortLinker:
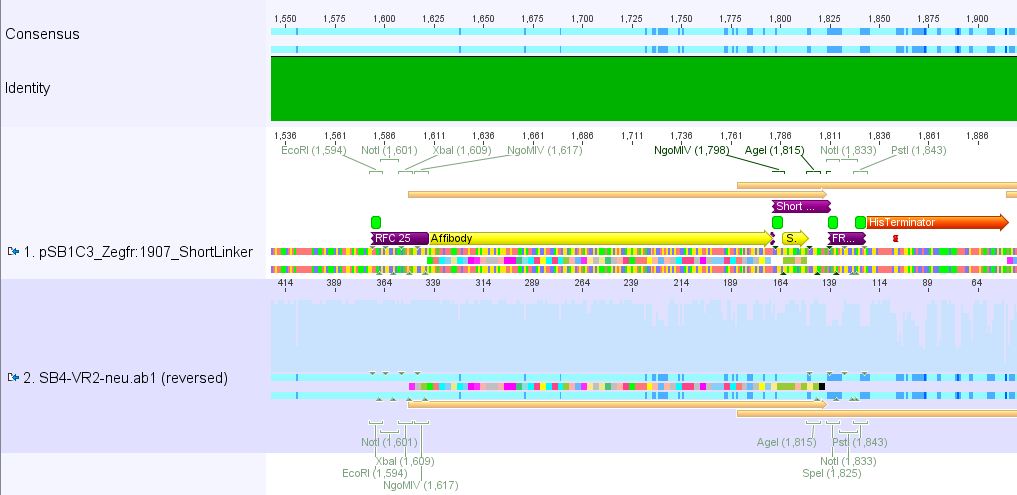
These constructs will be cloned into pCerulean for N-terminal fusion to VP2.
go to the top ;)
Trafo evalutaion and Inoculation
Investigator: Kira
The plate with transformed CD-biobrick contains around 40 colonies. 4 of them will be inoculated into LB and incubated @37 C over-night.
go to the top ;)
Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273)
Investigator: Patrick
The two Ligations were performed according to the standard protocol:
1 µl T4 DNA ligase, 1 µl 10x Buffer, 4,6 µl vector (P273), 3,34 µl Insert (P276, upper and lower band).
Incubation time: 50 minutes.
During the transformation i forgot to put the cells into the 37°C shaker so i incubated them afterwards again for 45 minutes at 37 °C on a shaker.
The mutual conles will be picked tomorrow to inoculate 10 ml DYT following a mini-prep on 03.09.2010
Transfection, Seeding AAV293 and HT1080
Investigator: Kerstin
- Transfection: 10 plates, same treatment (10µg of RC (P357), pHelper (P356)and mVenus (P263), following the standard protocol)
- Seeding HT1080 for transduction with virus from 21.08.2010 (10µg of RC, pHelper, TKGMK; pH=7,10)
- Seeding AAV293 for transfection with plasmids without HgH or beta-globin (verifying functionality)
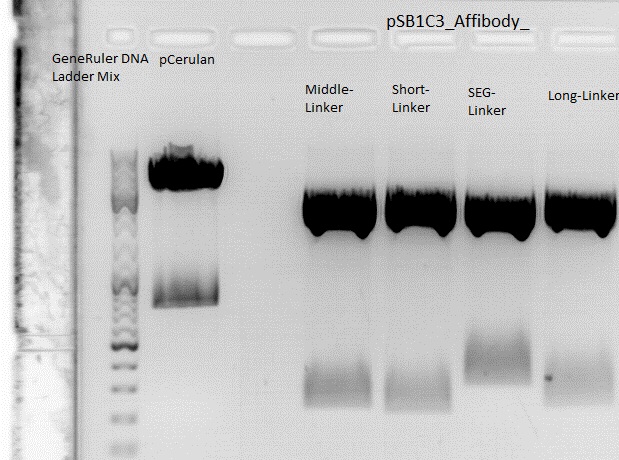
Cloning of ZEGFR:1907_Middle-Linker,ZEGFR:1907_Short-Linker, ZEGFR:1907_SEG-Linker and ZEGFR:1907_Long-Linker into pCerulean
Investigator: Stefan
Comment:
Digestion:
| components | Vector (P273) | ZEGFR:1907_Middle-Linker (P290) | ZEGFR:1907_Short-Linker (P292) | ZEGFR:1907_SEG-Linker (P296) | ZEGFR:1907_Long-Linker (P298) |
| DNA | 3,8 | 8,8 | 10,2 | 9,2 | 8,7 |
| BSA (10x) | 2 | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 |
| XbaI | 1 | 1 | 1 | 1 | 1 |
| PstI | 1 | 1 | 1 | 1 | 1 |
| H2O | 10,2 | 5,2 | 3,8 | 4,8 | 5,3 |
| Total volume | 20 | 20 | 20 | 20 | 20 |
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 115 Volt, running time: 50 minutes
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- P273: c= 3,1 ng/µl
- P290: c= 3,2 ng/µl
- P292: c= 1,8 ng/µl
- P296: c= 3,0 ng/µl
- P298: c= 3,2 ng/µl
T4 Ligation:
The Ligation was performed as following:
Comment: No sufficent vector concentration, therefore 5 µl of vector was used for every approach.
- Vector Volume: 5 µl
- Insert Volume: 3 µl
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
Incubating for 40 minutes.
Transformation:
Trafo was performed according to the standard protocol (BL21). The cells were plated on a agar plate with chloramphenicol
108. labday 02.09.2010
Sequencing of loop insertions BioBricks: 2nd round
Investigator: Achim, Anna
We analyzed the new sequencing results and found two correct sequences:
- 587 BAP clone 2.3
- 453 RGD clone 4.3
The other samples didn't contain any inserts or had wrong insertions. Apparently, the vector tends to religate easily. A possible reason for the religation of the incompatible ends is the overnight ligation at 18°C. We sent preps of the remaining 7 ligations for sequencing and will prep new clones or repeat the ligation tomorrow, depending on the new sequences.
Comment: Clones 1.4, 6.4, 9.4, 11.4, 12.4, 13.4, 14.4 were sent for sequencing. For sequencing results go to labday 03.09.10.
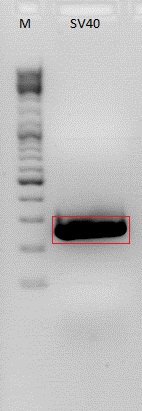
. Mini-preps of pSB1C3_SV40 and pCerulean_VP1_NLS
Investigator: Bea (Chris L)
Mini-Prep was performed according to the standard protocol
Construct were digested with XbaI and PstI for 45 minutes at 37°C. The loading plan on the 1% agarose gel looks like this:
_M_ ___ _363_ ___ _364_ _365_ _366_ _367_ _368_ _369_ _370_ ___
Results: P365 was sent for sequencing because test digestion looks goos. In contrast to the SV40 appraoch. This needs to be repeated tomorrow.
- Plasmid: pCerulean_VP1up_NLS
- Plasmid number: P365
- Tube name: SB1
- Primer used: CMV-F
Mini-Prep and test digestion of pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker
Investigator: Hanna
Comment: Zegfr:1907 (Affibody) and the His-Tag which was coupled to the middle linker were cloned into pCerulean. 3 clones of each construct were picked yesterday.
Plasmid Mini-Prep
- new vector name: pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker
Glycerol Stocks
1. pCerulean_Zegfr:1907
| Clone 1 | Clone 2 | Clone 3 | |
| Bacteria strain | XL1b | XL1b | XL1b |
| Plasmidname | pCerulean_Zegfr:1907 | pCerulean_Zegfr:1907 | pCerulean_Zegfr:1907 |
| Date | 2.9.10 | 2.9.10 | 2.9.10 |
| given glycerol-stock no. | B275 | B276 | B277 |
| given plasmid no. | P371 | P372 | P373 |
2. pCerulean_6xHis_MiddleLinker
| Clone 1 | Clone 2 | Clone 3 | |
| Bacteria strain | XL1b | XL1b | XL1b |
| Plasmidname | pCerulean_6xHis_MiddleLinker | pCerulean_6xHis_MiddleLinker | pCerulean_6xHis_MiddleLinker |
| Date | 2.9.10 | 2.9.10 | 2.9.10 |
| given glycerol-stock no. | B278 | B279 | B280 |
| given plasmid no. | P374 | P375 | P376 |
Test digestion
- buffer used: 4; Restriction-enzymes used: Enzyme 1 PstI-HF ; Enzyme 2 EcoRI-HF
- Plasmid
- Given Plasmid-Number: 371; DNA concentration: 381.3 ng/µL;
- Given Plasmid-Number: 372; DNA concentration: 377.5 ng/µL;
- Given Plasmid-Number: 373; DNA concentration: 409.7 ng/µL;
- Given Plasmid-Number: 374; DNA concentration: 404.2 ng/µL;
- Given Plasmid-Number: 375; DNA concentration: 366.1 ng/µL;
- Given Plasmid-Number: 376; DNA concentration: 375.0 ng/µL;
Comments::)
Pipetting scheme for each test digestion:
| Components | Volume/µL |
| DNA | 2.7 |
| BSA (10x) | 1 |
| Buffer no. 4 (10x) | 1 |
| EcoRI-HF | 0.5 |
| PstI-HF | 0.5 |
| H2O | 4.3 |
| Total volume | 10 |
- Incubation: 55 minutes
Agarose-Gel:
0.875 g Agarose, 50 mL TAE (1.75 %), 3 µL EthBr, at 115 Volt, running time: 35 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size |
|---|---|---|---|
| pCerulean_Zegfr:1907 clone 1 | 10 µl | 2 µl | 231 bp |
| pCerulean_Zegfr:1907 clone 2 | 10 µl | 2 µl | 231 bp |
| pCerulean_Zegfr:1907 clone 3 | 10 µl | 2 µl | 231 bp |
| pCerulean_6xHis_MiddleLinker clone 1 | 10 µl | 2 µl | 105 bp |
| pCerulean_6xHis_MiddleLinker clone 2 | 10 µl | 2 µl | 105 bp |
| pCerulean_6xHis_MiddleLinker clone 3 | 10 µl | 2 µl | 105 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample P371 /µl | Sample P372 /µl | Sample P373 /µl | Sample P374 /µl | Sample P375 /µl | Sample P376 /µl | |
|---|---|---|---|---|---|---|---|
| Lane | 5 | 12 | 12 | 12 | 12 | 12 | 12 |
Comments: Test digestion looked well:
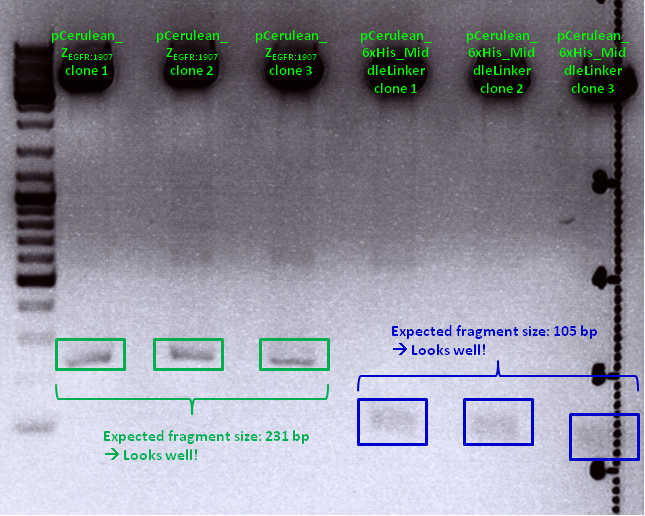
Clone 1 of each construct were sent for sequencing (P371 and P374 = HW1 and HW2). Used primer: GATC_std_CMV-F.
Midi-Prep of pSB1C3_lITR_CMV_mVenus_hGH_rITR clone1 & pSB1C3_lITR_CMV_beta-globin_mVenus_rITR clone1
Investigators: Chris W.
Midi-Preps of pSB1C3_lITR_CMV_mVenus_hGH_rITR clone1=P377 and pSB1C3_lITR_CMV_beta-globin_mVenus_rITR clone1=P378
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P377 | P378 |
| concentration (ng/µl) | 325,04 | 561,15 |
Mini-Prep and test digestion of the CD biobricks
Investigator: Kira
Wh did you digest with eco and ndeI?? is there a spechía reasonn for it?? if the coonstruct was in the rfc standard could digest it with the igem standard enzymes??
| components | sample /µl |
| DNA | 2,5 |
| BSA (100x) | 0 |
| Buffer ___4_ (10x) | 1,5 |
| Enzyme NdeI (no.Lab:___) | 0,5 |
| Enzyme EcoRI (no.Lab:___) | 0,5 |
| H2O | 10 |
| Total volume (e.g. 15,20,25,30 µl) | |15 |
Sample 1 was sent for sequencing
109. labday 03.09.2010
Miniprep and test digestion of pSB1C3_lITR_pTERT_ßglobin_mVenus_hGH_rITR
Investigator: Achim
Comment: The test digestion did not show the expected fragments of 2000 and 2500 bp. the photo was exposed too long to see distinct bands. Digestion will be repeated.
Sequencing results
Investigator: Hanna
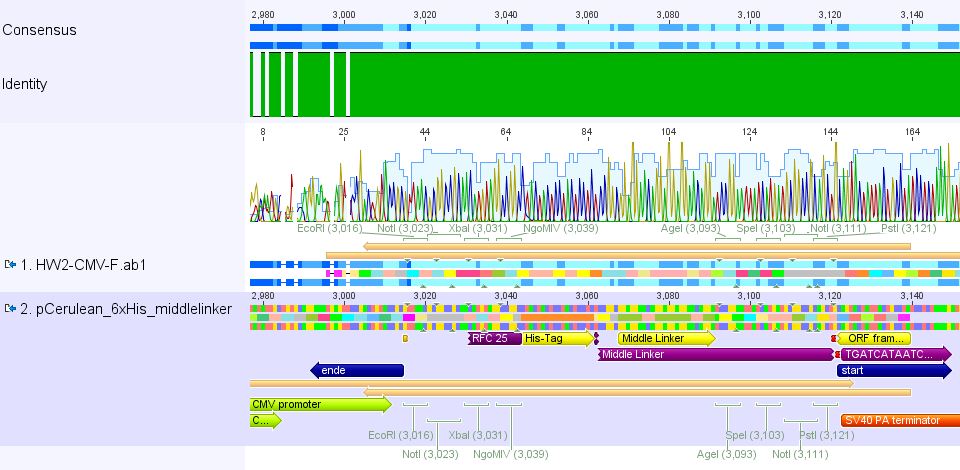
Comment: pCerulean_Zegfr:1907 and pCerulean_6xHis_MiddleLinker were cloned and sent for sequencing yesterday.
1. pCerulean_Zegfr:1907: Sequencing looked well.
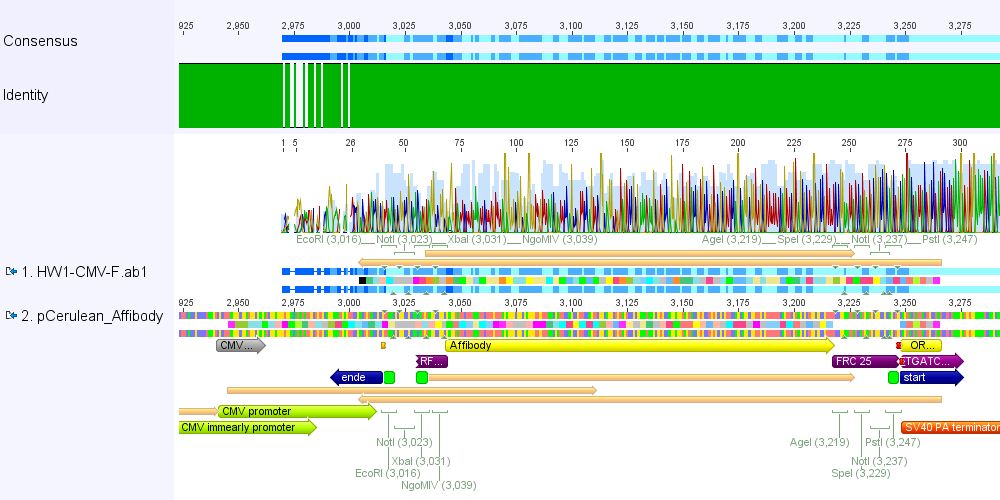
2. pCerulean_6xHis_MiddleLinker: Sequencing looked also well.
Cloning of pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker
Investigator: Hanna
Comment: For N-terminal fusion to VP2, the Affibody - fused to different kinds of linkers - will be cloned into the expression plasmid pCerulean. For imaging CFP, which was fused to the middle linker, will be cloned into pCerulean.
Practical Cloning:
- plasmid:
- Vector: name: pCerulean number: P273
- Insert: name: pSB1C3_Zegfr:1907_LongLinker (P298), pSB1C3_Zegfr:1907_MiddleLinker (P290), pSB1C3_Zegfr:1907_SEG (P296), pSB1C3_Zegfr:1907_ShortLinker (P292), pSB1C3_CFP_MiddleLinker (P276)
- new vector name: pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 EcoRI-HF ; Enzyme 2 PstI-HF
- DNA concentration (vector): 419.5 ng/µL ; DNA concentration (insert): P298 229.7 ng/µL, P290 227.4 ng/µl; P296 218.7 ng/µL; P292 196.6 ng/µL; P276 207.7 ng/µL
Comments: No.
Digestion
| components | volume of pCerulean /µl | volume of Zegfr:1907_MiddleLinker /µl | volume of CFP_MiddleLinker /µL |
| DNA | 8.3 | 7.6 | 4.8 |
| BSA (10x) | - | - | - |
| Buffer 4 (10x) | 2 | 2 | 2 |
| Enzyme 1 EcoRI-HF | 1 | 1 | 1 |
| Enzyme 2 PstI-HF | 1 | 1 | 1 |
| H2O | 7.7 | 8.4 | 11.2 |
| Total volume | 20 | 20 | 20 |
- Incubation: 1.5 h
Agarose-Gel:
0.8 g Agarose, 50 TAE (1.5 %), 5 µL EthBr, at 70 Volt, running time: ~ 1 hour
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 |
|---|---|---|---|
| P273 | 20 µl | 4 µl | 3922 bp |
| P298 | 20 µl | 4 µl | 273 bp |
| P290 | 20 µl | 4 µl | 261 bp |
| P296 | 20 µl | 4 µl | 345 bp |
| P292 | 20 µl | 4 µl | 249 bp |
| P276 | 20 µl | 4 µl | 801 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample P273 /µl | Sample P276 /µl | Sample P290 /µl | Sample P292 /µl | Sample P296 /µl | Sample P298 /µl | |
|---|---|---|---|---|---|---|---|
| Lane | 5 | 24 | 24 | 24 | 24 | 24 | 24 |
Gel extraction
Gel measurement:
| Sample | Volume | Concentration |
| P273 | 20 | 115.45 ng/µL |
| P276 | 20 | 9.91 ng/µL |
| P298 | 20 | 5.07 ng/µL |
| P296 | 20 | 9.03 ng/µL |
| P290 | 20 | 10.31 ng/µL |
| P292 | 20 | 20.78 ng/µL |
Ligation
1. PCerulean_Zegfr:1907_LongLinker
| P298 | P273 | |
| Volume/µl | 6.61 | 1.39 |
2. PCerulean_Zegfr:1907_MiddleLinker
| P290 | P273 | |
| Volume/µl | 5.53 | 2.47 |
3. PCerulean_Zegfr:1907_SEG
| P296 | P273 | |
| Volume/µl | 6.17 | 1.83 |
4. PCerulean_Zegfr:1907_ShortLinker
| P292 | P273 | |
| Volume/µl | 4.11 | 3.89 |
5. PCerulean_CFP_MiddleLinker
| P276 | P273 | |
| Volume/µl | 7.02 | 0.98 |
Trafo
Trafo was performed following the standard protocol. Used cells: XL1b, DNA amount: 2.5 µL of each ligation reaction. Plates were stored @ 37°C room over night.
Harvest viral particles, Seeding HT1080 cells
Investigator: Adrian, Kerstin
- Harvest viral particles of Transfection from 31.08.2010 (six approaches: 10µg of RC (2x P326 and 2x P325 and 2x Stratagene), pHelper and GOI (P262))
- Seeding HT1080 cells
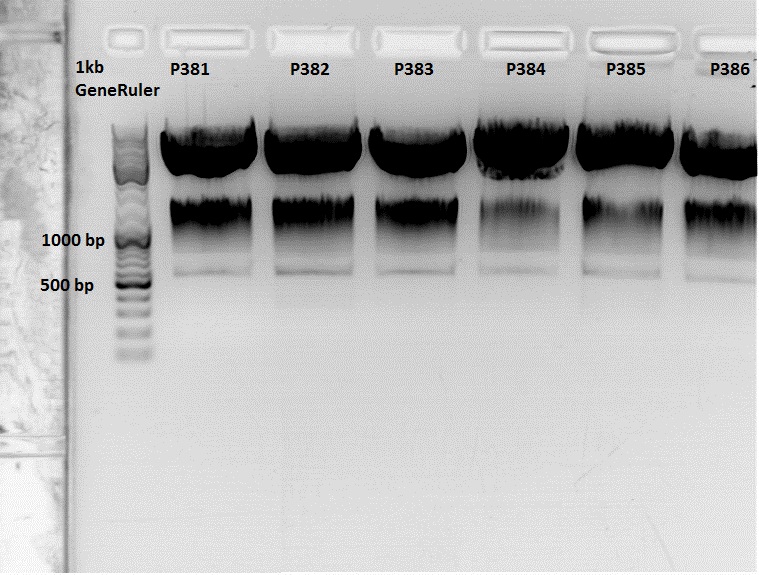
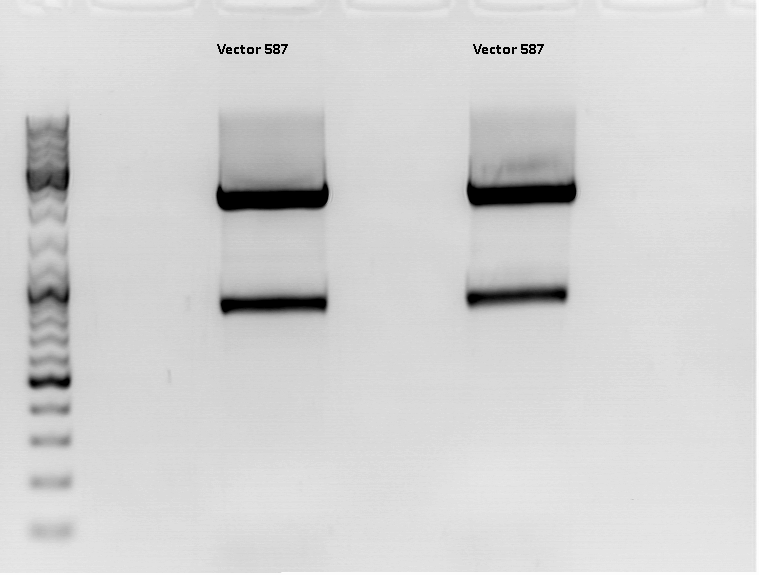
Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273)
Investigator: Patrick
The Miniprep was performed according to the standard protocol.
Labelling:
- P381: pCerulean_middlelinker clone 1 upper band : 526,0 ng/µl
- P382: pCerulean_middlelinker clone 2 upper band : 522,0 ng/µl
- P383: pCerulean_middlelinker clone 3 upper band : 463,0 ng/µl
- P384: pCerulean_middlelinker clone 1 lower band : 465,0 ng/µl
- P385: pCerulean_middlelinker clone 2 lower band : 500,0 ng/µl
- P386: pCerulean_middlelinker clone 3 lower band : 524,0 ng/µl
Testdigestion: 7 µl DNA sample, 1 µl Buffer 4 (10x), 1 µl BSA, 0,5 µl Xba, 0,5 µl PstI-HF
Expected size of the fragments: 786 & 2053bp
There seems to be something wrong.
Sent for sequencing: P381 & P385
Labelling: P381-CMV_rev & P385-CMV_rev
Used primer: O21 : CMV_reverse_qPCR
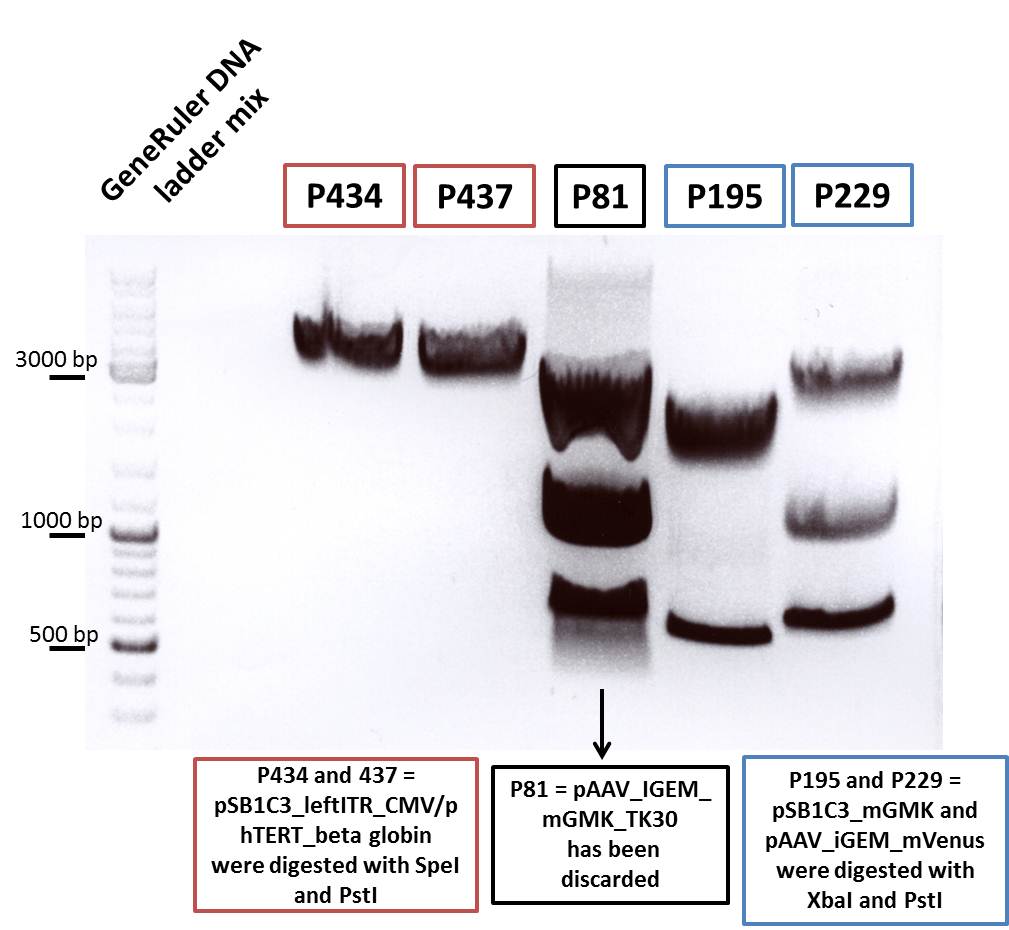
Cloning of pSB1C3_leftITR_pTert and pSB1C3_mGMK with pSB1C3_leftITR_CMV
Investigator: Chris L.
- Vector: name: pSB1C3_leftITR_pTERT clone 1 P256 c=179,2 ng/µl
- Vector: name: pSB1C3_leftITR_CMV P188 c=231,6 ng/µl
- Insert: name: pSB1C3_mGMK P195 c=258,6 ng/µl
- new vector name: pSB1C3_leftITR_CMV_GMK
- new vector name: pSB1C3_leftITR_pTERT_GMK
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 PstI ; Enzyme 2 SpeI ; Enzyme 3 XbaI
| components | P195 | P256 /µl | P188 /µl |
| DNA | 7,7 | 11,2 | 8,6 |
| BSA (10x) | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 |
| Enzyme SpeI | 1 | 1 | 0 |
| Enzyme XbaI | 0 | 0 | 1 |
| Enzyme PstI | 1 | 1 | 1 |
| H2O | 6,3 | 2,8 | 5,4 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 | 20 |
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 130 Volt, running time:45
Results:
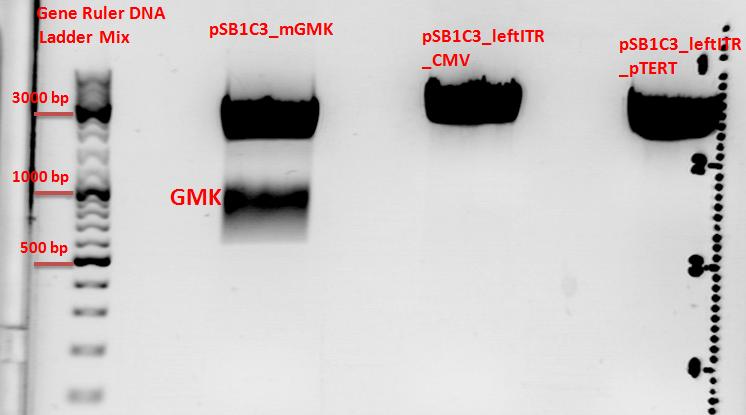
- c(Insert)= 15,37 ng/µl; size: 627 bp
- c(Vector)= 50,84 ng/µl; size: 2869 bp
- c(Vector)= 75,64 ng/µl; size: 2672 bp
- Ligation of PCR products and vector:
For the Ligation 1µl T4 buffer (10x) and 1µl T4 ligase were used. Incubation time: 45 min
| pSB1C3_leftITR_CMV + GMK /µl | pSB1C3_leftITR_pTERT + GMK | |
| Vector | 5,48 | 6,21 |
| Insert | 2,52 | 1,79 |
- Transformation:
The transformation was done following the standard protocol using XL1 blue cells.
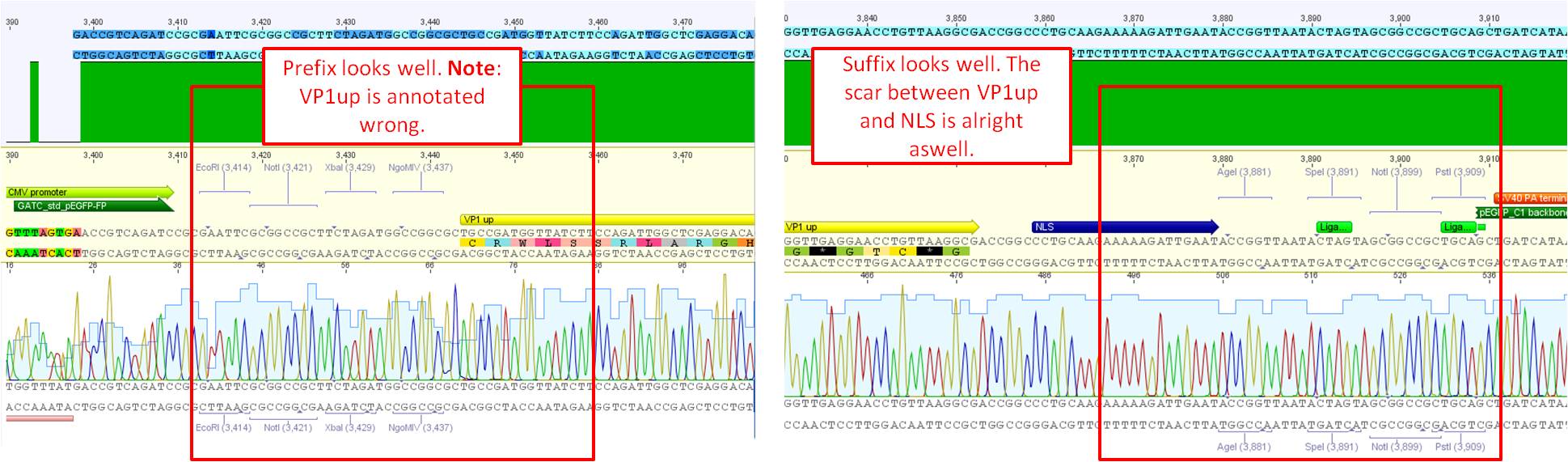
Sequence analysis of pCerulean_VP1up
Investigator: Bea
The assembled pCerulean_VP1up which serves as the first construct in the VP1 insertion cloning assembly was sent for sequencing.
- Plasmid used: P365
- Primer used: CMV-F
- Tube name: SB1
- Folder (Geneious): N-terminal Targeting --> pCerulean_VP1up_NLS
pCerulean_VP1up_NLS_Targeting molecule
Investigator: Bea
Comment: For preparing the next step in the VP1 insertion, different targeting molecules will be fused to the construct pCerulean_VP1up_NLS.
- The first construct is the Affibody ZEGF-R:1907 which was already cloned into the standard iGEM plasmid and can be sued for target tumor cells overexpressing the EGF receptor.
- The second construct is the mVenus (Yellow fluorescent protein) which can be used for imaging experiments
- Another motif will be the His-tag. Since the His-tag is very small, the oligos will be hybridized and directly ligated into the pCerulean backbone.
Digestion of vector:
- Plasmid used: pCerulean_VP1up_NLS (P365) c=470 ng/µL
- Add BSA
- AgeI-HF and SpeI-HF were used
Protocol of the digestion of the vector:
| Components | vpCerulean_VP1up_NLS/µL |
| DNA | 3 |
| BSA (100x) | 2,5 |
| Buffer no. 4 (10x) | 2,5 |
| Enzyme 1 AgeI | 1 |
| Enzyme 2 SpeI HF | 1 |
| H2O | 15 |
| Total volume | 25 |
- Incubation of the sample at 37°C
- Incubation for 120 minutes
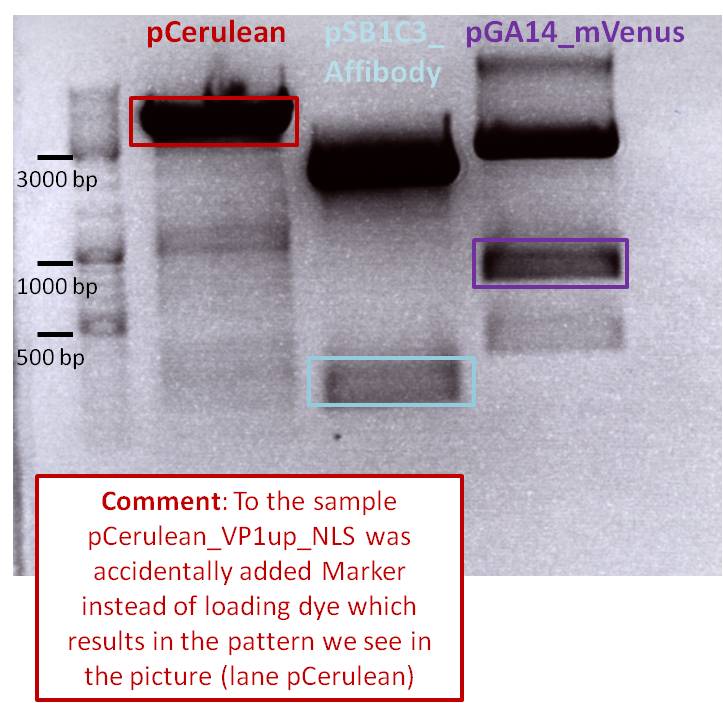
The Targeting molecules were digested with different enzmyes than the vector for providing the ability to assemble the different constructs together without creating new restriction site but to delete them. This works by fusing NgoMIv and AgeI together.
Digestion of the targeting molecules
- Plasmids used:
- pSB1C3_zEGF-R:1907 (P284) c=146 ng/µL
- pGA14_mVenus(P60) c=280 ng/µL
- Add BSA
- NgoMIV and SpeI-HF were used
| Components | vZEGFR (P284)/µL | vpGA_mVenus (P60)/µL |
| DNA | 13 µl | 7 µl |
| BSA (10x) | 2,5 µl | 2,5 µl |
| Buffer no. 4 (10x) | 2,5 µl | 2,5 µl |
| Enzyme 1 NgoMIV- | 1,0 µl | 1,0 µl |
| Enzyme 2 AgeI HF | 1,0 µl | 1,0 µl |
| H2O | 15 µl | 15 µl |
| Total volume | 25 | 25 |
- Incubation of the sample at 37°C
- Incubation for 90 minutes
After incubation the samples were loaded on a 1% agarose gel. The obtained gel (after running the gel for 15 minutes) can be seen below.
Results: The boxes mark fragments which ere cut out of the gel. The left lane (pCerulean) looks a bit strange, because Marker was added after the digestion reaction instead of Loading dye.
Parallel, for assembling the His-Tag to the construct pCerulean_VP1up_NLS, oligos for the His-Tag in the RFC25 standard (provided by Gerrit) were hybridized and can directly be ligated in the digested vector. This strategy was used because of the smal size of the His-Tag which cannot be separated easily by the agarose gel.
Hybridization of the His-Oligos
- 10µL oligo1 (1:10)
- 10µL oligo2 (1:10)
- 4µL 100mM Tris-Cl pH 8
- 10µL 5mM MgCl2
- 8µL H2O
The used programm for the hybridization:
- 99°C 7´
- 99°C 1´
- -1°C R=0,3°/s
- Goto 2 rep 74
- Hold 4°C
After the gel extraction of a T4 ligation was performed.
Affibody ZEGF-R:1907:
vCerulean_VP1up_NLS = 5,82µL
vZEGF-R:1907 = 2,18µL
mVenus Z:
vCerulean_VP1up_NLS = 4,04µL
vmVenus = 3,96 µL
6xHisTag:
vCerulean_VP1up_NLS = 0,2µL
v6xHisTag = 7,98µL
Next steps: Picking clones of the trafo plates (containing Kanamycin) and perform the Mini-Prep. The obtaining constructs finally will be fused to the VP2/3 protein which results in the final construct for the VP1 targeting approach.
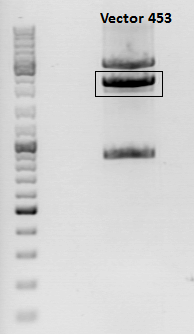
3rd Repetition of Mini-Preps and test digestion for Loop insertion BioBricks
Investigator: Achim, Anna
Sequencing results of Loop insertion BioBricks from 02.09.10; the following clones looked well:
- pSB1C3_587_KO_RGD_clone6.4
- PSB1C3_587_KO_His_clone9.4
Update: 9 of the 14 ViralBricks looked well (sequencing results from 01. - 03.09.). Two new clones of the remaining constructs were prepared and test digested.
Test digestion:
| components | Volume for each sample /µl |
| DNA | 10 |
| BSA (10x) | 1,5 |
| Buffer 4 (10x) | 1,5 |
| Enzyme EcoRI HF | 0,5 |
| Enzyme NotI HF | 0,5 |
| H2O | 1 |
| Total volume /µl | 15 |
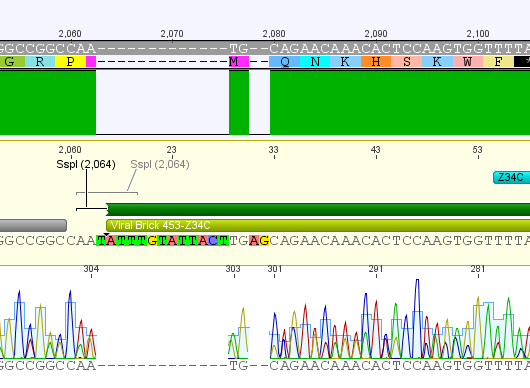
| Components | 453 BAP | 453 Z34C | 587 Z34C | 587 KO Z34C | 587 KO Z34C SPACER |
| Clone | 1.5 + 1.6 | 11.5 + 11.6 | 12.5 + 12.6 | 13.5 + 13.6 | 14.5 + 14.6 |
Incubation time: 1 h, Incubation temperature: 37°
Preparation of gel:
1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 45 minutes
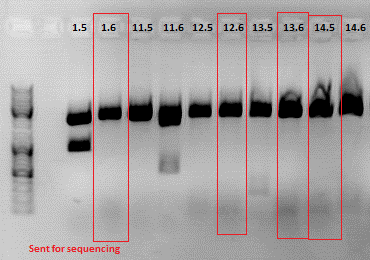
Expected fragment sizes /bp:
pSB1C3: 2051 bp
BAP: ~150
Z34C: ~ 200
Comment: The following clones were sent for sequencing: 1.6, 12.6 ,13.6 and 14.5.
To do: Retrafo of clone6.4 and clone9.4 and preparation of glycerol stocks.
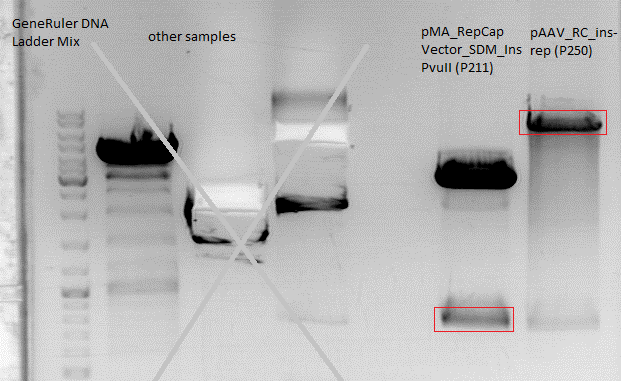
Cloning of Cap into pAAV_RC_ins-rep: preparation for SDM
Investigator: Stefan
Comment: Cloning Cap into pAAV still does not work as planned. As another approach Cap can be cut using BsiWI and XcmI and performing a SDM in the part of Cap not cloned into pAAV. Primers are ordered and should arrive on monday to proceed.
Digestion:
| components | pMA_RepCap Vector_SDM_InsPvuII (P211) | pAAV_RC_ins-rep (P250) |
| DNA | 7,9 | 2,1 |
| Buffer 2 (10x) | 2 | 2 |
| XcmI | 1 | 1 |
| BsiWI | 1 | 1 |
| H2O | 8,1 | 13,9 |
| Total volume | 20 | 20 |
Comment:Digestion was performed using two steps, first incubating for 1 hour at 37 °C, afterwards for 1,5 hours at 55 °C.
Gel:
0,5 g Agarose,50 ml TAE (1%), 5 µl EtBr , at 115 Volt, running time: 50 minutes
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- pMA_RepCap Vector_SDM_InsPvuII (P211): c= 6,26 ng/µl
- pAAV_RC_ins-rep (P250): c= 13,72 ng/µl
Quick Ligation:
Comment: XcmI produces only 1 base overhang, therefore Quickligase was used for ligation and incubation time increased.
The Ligation was performed as following:
- Vector Volume: 4,59 µl
- Insert Volume: 4,41 µl
- 10 µl QuickLigase buffer (2x)
- 9 µl (Vector + Insert) mix
- 1 µl QuickLigase
Incubating for 60 minutes.
Transformation:
Trafo was performed according to the standard protocol (XL1b). The cells were plated on a agar plate with ampicilin.
110. labday 04.09.2010
Continuation: Cloning CFP_middlelinker (from pSB1C3_CFP_middlelinker, P276) into pCerulean (P273): Sequencing results of P381 and P385
Investigator:Patrick
Sequencing results of P381 & P385 Labelling: P381-CMV_rev & P385-CMV_rev: A wrong primer was used so there are no relevant sequence data yet. On sunday i will sent these clones again for sequencing. Primer: GATC_std_CMV-F
Hiermit verspreche ich dass es noch jede Menge Kopfkratzer, Laboraufsichtswichtel und pinke Panter geben wird (Patrick)
Harvest viral particles, Transduction
Investigator:Kerstin, Adrian
- Harvest viral paricles from one plate of transfection from 01.09.2010 (10 plates, same treatment (10µg of each plasmid P357, P263, P356))
- Transduction:
- 1x 6-well: 0,5ml of virus from 01.09.2010 (testing R/C P326)
- 1x 6-well: 0,5ml of virus from 01.09.2010 (testing R/C P325)
- 4x 6-well: 10x 0,5ml and 4x 1ml of virus from 01.09.2010 (stratagene R/C)
- 4x 6-well: 10x 0,5ml and 4x 1ml of virus from 04.09.2010 (10 plates same treatment)
- 6x 6-well: 0,5ml of virus from 21.08.2010 (TKGMK; 18x pH 7,10 and 6x pH 7,12)
Repetition of Hybridisation and Cloning for Loop insertion BioBricks
Investigator: Achim, Anna
- Samples:
1: 453_BAP
11: 453_Z34C
12: 587_Z34C
13: 587_KO_Z34C
14: 587_KO_Z34C_Spacer
Comment: New approaches of the Hybridisation and Fill-in reactions were done because it didn't work the first time. A possible reason is that the klenow enzyme wasn't inactivated. In addition, the conditions for ligation will be improved. Also the ligation of 453_BAP was done again.
Digestion:
| components | Volume Vector 587 | Volume Vector 453 | Sample 11 | Sample 12 | Sample 13 | Sample 14 |
| DNA | 3,7 | 3,7 | 6 | 6 | 6 | 6 |
| BSA (10x) | - | - | - | - | - | - |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 | 2 |
| Enzyme 1° | Bam | Ssp | Ssp | Bam | Bam | Bam |
| Enzyme 2° | Pvu | Sal | Sal | Pvu | Pvu | Pvu |
| H2O | 12,3 | 12,3 | 10 | 10 | 10 | 10 |
| Total volume /µl | 20 |
° 1 µl each
Preparation of gel:
1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 50 minutes
Repetition: Prepatation for SDM: cloning of Cap into pAAV
Investigator: Stefan
Comment: On yesterday's plates nothing grew. Because gelex and ligation products were stored in coldroom, new ligation and trafo approaches were performed.
Ligation approaches:
- ligation with Quick Ligase (as performed yesterday)
- ligation with T4 Ligase
Transformation approaches:
- Quick Ligase (approach from yesterday):
- 2 µl of ligation product
- 4 µl of ligation product
- Quick Ligase (new approach):
- 2 µl of ligation product
- 4 µl of ligation product
- T4 Ligase:
- 2 µl of ligation product
- 4 µl of ligation product
Transformation:
Trafo of each approach was performed according to the standard protocol (XL1b). The cells were plated on a agar plate with ampicilin.
Ligation and trafo of the modified Cap insert , the Viral Bricks 1, 11, 12, 13 and 14
Investigator: Volker
Ligation
| Construct | Vector (µl) | Insert(µl) |
| ViralBrick 1 in 453 | 7.91 | 0.09 |
| ViralBrick 11 in 453 | 6.17 | 1.83 |
| ViralBrick 12 in 587 | 6.15 | 1.85 |
| ViralBrick 13 in 587 | 5.3 | 2.7 |
| ViralBrick 14 in 587 | 4.46 | 3.54 |
| p211 in pAAV-RC | 5.52 | 3.54 |
| p211 in pAAV-RC | 5.52 | 3.54 |
Preparation of PBS Buffer
Investigator: Volker
Two liters of PBS buffer were prepared for the column purification of the viral particles. The following protocol was used:
- dissolve the following in 800ml ddH2O:
- 8g of NaCl
- 0.2g of KCl
- 1.44g of Na2HP0$ (1.69 because of the crystal water in the reagent)
- 0.24 g of KH2PO4
- adjust to pH 7.4
- adjust to 1000ml
- steril filtrate the solution
111. labday 05.09.2010
Mini-prep of glycerol stock pMA_RepCap Vector_SDM_InsPvuII clone 1 (B182)
Investigator: Stefan
Glycerol stock: Because glycerol stock B182 was not mixed before putting into -80 °C freezer the cells died. Therefore a new glycerol stock was prepared and labled B302.
Mini-Prep was performed according to the standard protocol. Two preps were prepared:
Picking clones of pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker
Investigator: Hanna
2 clones of each construct (1. pCerulean_Zegfr:1907_ShortLinker, 2. pCerulean_Zegfr:1907_MiddleLinker, 3. pCerulean_Zegfr:1907_LongLinker, 4. pCerulean_Zegfr:1907_SEG and 5. pCerulean_CFP_MiddleLinker) were picked.
To do tomorrow (6.9.): Mini-Preps and Test digestion (with EcoRI and PstI).
112. labday 06.09.2010
Minipreps & Test PCR of pTert Biobrick assembly
Investigator: Achim
- After unsucessful test digestion with EcoRI and PstI I tried out a test PCR with Igem Primers (same Mastermix as for loop inserts)
- Elongation time was set to 2.35 min because the amplified fragment should be about 2.5 kbp big.
- Result:
- Next steps: Another test digestion tomorrow, if that doesn't work out I'll repeat the ligation.
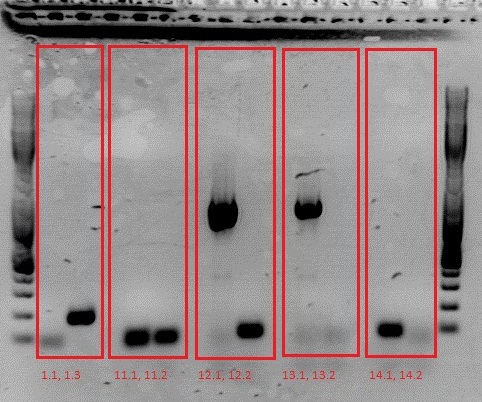
Minipreps & Test PCR of Loop inserts 1, 11, 12 ,13, 14
Investigator: Achim & Anna
- Samples:
1: 453_BAP
11: 453_Z34C
12: 587_Z34C
13: 587_KO_Z34C
14: 587_KO_Z34C_Spacer
- Minipreps of 1.1, 1.3, 11.1, 11.2, 12.1, 12.2, 13.1, 13.2, 14.1, 14.2
- Clones 1.4, 11.3, 11.4, 12.3, 12.4, 13.3, 13.4, 14.3, 14.4 were pelleted and stored away in case the first two are incorrect.
- In the hope to confirm our small inserts, A test PCR using Igem standard primers was carried out:
PCR-reaction:
| Volume / µl | ingredients |
| 2,5 | Taq Buffer |
| x | template (~0.5 ng) |
| 2,5 | forward Igem primer (1:10) |
| 2,5 | reverse Igem primer (1:10) |
| 0,5 | dNTP |
| filled upt to 25 | H2O |
| 0,5 | Taq Polymerase (1.25) |
PCR program:
| Rounds | temperature/ °C | Time |
| 1 | 95 | 30" |
| 95 | 25" | |
| 25 | 61 | 30" |
| 68 | 50" | |
| 1 | 68 | 5" |
Results:
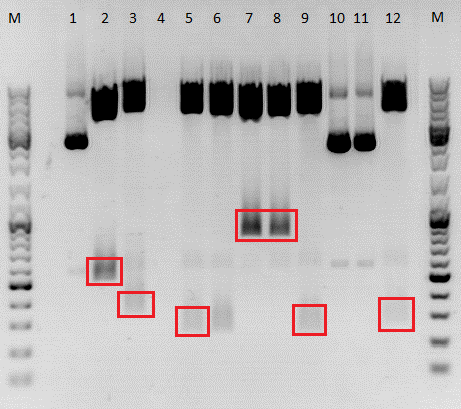
Mini-Preps and test digestion of Linker BioBricks
Investigator: Anna
Comment: Mini-preps of all clones were prepared. They have not been added to the excel sheets yet, but were stored in the freezer -> Samples of the usable and sequenced constructs were added to the excel sheets and stored in the freezer (06.09.10).
| Samples | 1 | 2 | 3 | 4 | 5 | 6 |
| Insert | VP1up_NLS_His | VP1up_NLS_His | ZeGFP:1907_SEG | ZeGFP:1907_SEG | ZeGFP:1907_ML | ZeGFP:1907_ML |
| Clone | 1 | 2 | 1 | 2 | 1 | 2 |
| Concentration | 439,57 | 463,51 | 415,24 | 354,5 | 406,75 | 382,27 |
| 7 | 8 | 9 | 10 | 11 | 12 |
| CFP_ML | CFP_ML | ZeGFP:1907_SL | ZeGFP:1907_SL | ZeGFP:1907_LL | ZeGFP:1907_LL |
| 1 | 2 | 1 | 2 | 1 | 2 |
| 482,76 | 424,76 | 497,06 | 413,55 | 391,05 | 427,20 |
SL: Shortlinker
ML: Middlelinker
LL: Longlinker
Test digestion:
| components | Volume for each sample /µl |
| DNA | 1 |
| BSA (10x) | - |
| Buffer 4 (10x) | 1 |
| Enzyme EcoRI HF | 0,5 |
| Enzyme Pst HF | 0,5 |
| H2O | 7 |
| Total volume /µl | 10 |
Preparation of gel:
- Incubation time: 1 h, Incubation temperature: 37°
- 1 g Agarose, 100 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 45 minutes
- Expected fragment sizes:
VP1up_NLS_Histag: 550 bp
ZeGFP:1907_SEG: 344 bp
ZeGFP:1907_ML: 261 bp
CFP_ML: ~ 1000 bp
ZeGFP:1907_SL: 249 bp
ZeGFP:1907_LL: 237 bp
To do: Test digestion of VP1up_NLS_mVenus and Affibody. Number 2, 3, 5, 7, 9 and 12 will be sent for sequencing tomorrow. For sequencing results of VP1up_NLS_Histag go to labday 09.09.2010, for the linker BioBricks go to labday 08.09.2010.
Production of Ampicilin and Chloramphenicol
Investigator: Jessica
- 10ml ethanol (70%) containing 1g Amp in 60µl aliquots
- 10ml ethanol (70%) containing 0,25g Cm in 60 µl aliquots
Mini-prep and test digestion of pAAV_RC-CapIns_prepSDM
Investigator: Stefan
Mini-Prep was performed according to the standard protocol
Constructs were digested with XcmI and Acc65I for 70 minutes at 37°C
| components | pAAV_RC_CapIns_prepSDM clone 1-8 |
| DNA | 2 |
| Buffer 2 (10x) | 1 |
| BSA (10x) | 1 |
| XcmI | 0,5 |
| Acc65I | 0,5 |
| H2O | 5 |
| Total volume | 10 |
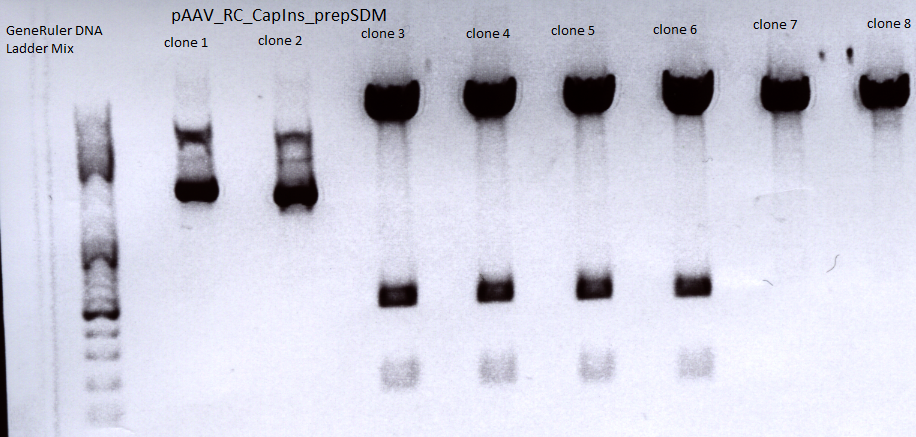
Sent for sequencing:
Clone 3 (P396) was sent for sequencing.
- Tube name: SB1
- Primer used: VP 4200 rev
Sent for sequencing
Investigator: Stefan
- Tube name: SB2
pAAV_RC_inserts clone1 (P326) was sent for sequencing using the following primers:
- VP1 primer for pKex rev
- SK
- Primer Cap 2800 for
- Primer Cap 2800 rev
- Primer Cap 3500 for
113. labday 07.09.2010
Sequencing results of new loop insertion ligations
Investigator: Achim
- Sequencing confirmed the 453 BAP ligation contains the correct sequence. 453 Z34C and 587 Z34C didn't contain inserts, 587 KO Z34C still contained the BLA placeholder.
Repetition of CD biobrick production
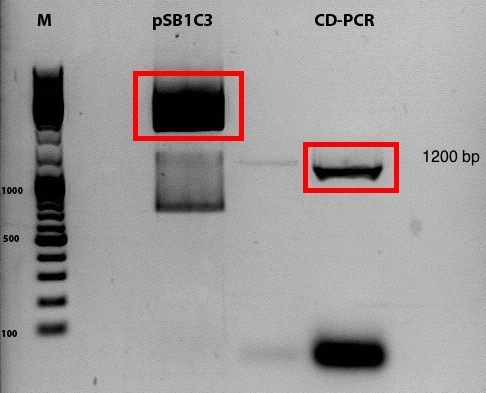
Comment: Despite the results of the test digestion, the samples, which were sent for sequencing do not contain the GOI. Thus the whole procedure will be repeated today.
Investigator: Kira
DNA sample was diluted 1:100
| Ingredients | CD sample |
| 5X Phusion HF buffer | 10 µl |
| 10 mM dNTP mix | 1µl |
| forward primer: O158 | 2,5µl |
| reverse primer: O159 | 2,5 µl |
| DNA Template | 0,5 µl |
| DMSO | 0 µl |
| Phusion Polymerase | 0,5 µl |
| H2O | 33µl |
| Total volume | 50 µl |
PCR program:
| Cycles | Temperature | Time |
| 98°C | 1 | |
| 8x | 98°C | 15" |
| 60°C | 25" | |
| 72°C | 60" | |
| 17x | 98°C | 15" |
| 65°C | 25" | |
| 72°C | 60" | |
| 1x | 72°C | 5' |
| Hold 4°C |
Digestion of plasmid backbone:
c (pSB1C3) = 151, 1 ng/ µl
| Components | vector Volume/µL |
| DNA 1 µg | 6,0 µl |
| BSA (100x) | 0,2 µl |
| Buffer no. 4 (10x) | 2,0 µl |
| Enzyme 1 XbaI | 0,5 µl |
| Enzyme 2 AgeI HF | 0,5 µl |
| H2O | 10,8 µl |
| Total volume | 20 |
incubation @ 37 C for approx. 2 h
1% agarose gel
Digestion of PCR product:
| Components | PCR product Volume/µL |
| DNA | 30,0 µl |
| BSA (100x) | 0,4 µl |
| Buffer no. 4 (10x) | 4,0 µl |
| Enzyme 1 XbaI | 1,5 µl |
| Enzyme 2 AgeI HF | 1,0 µl |
| H2O | 3,1 µl |
| Total volume | 40 |
incubation @ 37 C for approx. 2 h
Ligation
Quickligase was used
DNA-mix 9 ul (8,2 ul insert +0,8 ul insert)
Quickligase 1ul
Quickligase Buffer 10 ul
Incubation @ RT for 10'
Transformation
BL21 cells were used for transformation because XL1B cells have to be tested first. Transformation according to the standard protocol.
Comment: to figure out if, the digestion of the vector and the following ligation was successful, 2 plates have been prepared. One plate contains only the digestion vector, while the second contains the ligated sample.
Test digestion of pCerulean_VP1up_NLS_mVenus and pCerulean_VP1up_NLS_Affibody
Investigator: Anna
| Samples | Insert | Clone | Concentration [ng/µl] |
| 1 | VP1up_NLS_mVenus | 1 | 416,59 |
| 2 | VP1up_NLS_mVenus | 2 | 387,46 |
| 3 | VP1up_NLS_Affibody | 1 | 390,68 |
| 4 | VP1up_NLS_Affibody | 2 | 374,12 |
Test digestion:
| components | Volume for each sample /µl |
| DNA | 2 |
| BSA (10x) | - |
| Buffer 4 (10x) | 1 |
| Enzyme EcoRI HF | 0,5 |
| Enzyme Pst HF | 0,5 |
| H2O | 6 |
| Total volume /µl | 10 |
Preparation of gel:
- Incubation time: 2 h, Incubation temperature: 37°
- 0,5 g Agarose, 50 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 45 minutes
- Expected fragment sizes:
VP1up_NLS_mVenus: 1215 bp
VP1up_NLS_Affibody: ~ 700 bp
Comment: Samples 1 and 3 were sent for sequencing. For sequencing results go to labday 09.09.2010.
Reftrafo of pSB1A2_EGFP_HQ from distribution plate (Bba_I719015 plate 1 well 15P = T7GFPmut3bHQ)
Investigator: Jessica
P15 (T7GFPmut3bHQ) from plate 1 of the distribution plates from the iGEM headquarter was solved in 10 µl H2O
- two trafos was prepared with 2 µl DNA and XL1blue (lab)
- plate 1: 90µl DYT + 10µl mixed DNA-XL1b-solution
- plate 2: resupended pellet
Design and Ordering of GFP-Oligos
Investigator: Hanna
Comment: In order to perform single virus tracing experiments and in order to test the Affibody and the DARPin, we decided to use GFP which can be better visualized than CFP. Because we received GFP in the distribution kit we performed a trafo with the Bba_I719015. In addition to that we want to convert the GFP into the RFC25 standard - because for our purpose we want to be able to perform fusion approaches.
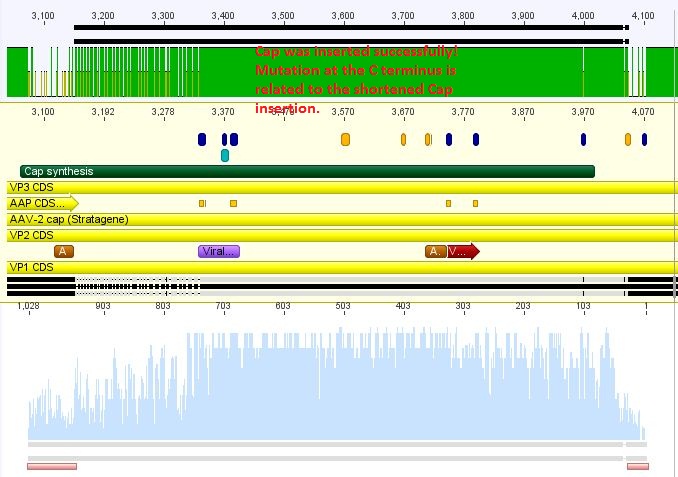
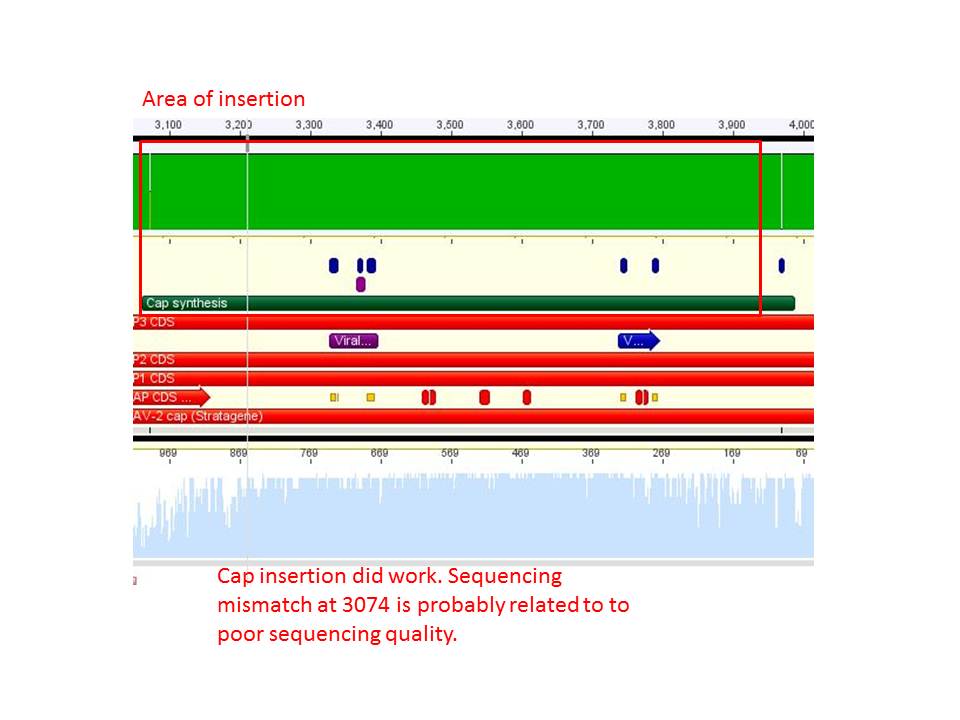
Sequence analysis of pAAV_RC_CapIns_prepSDM
Investigator: Stefan
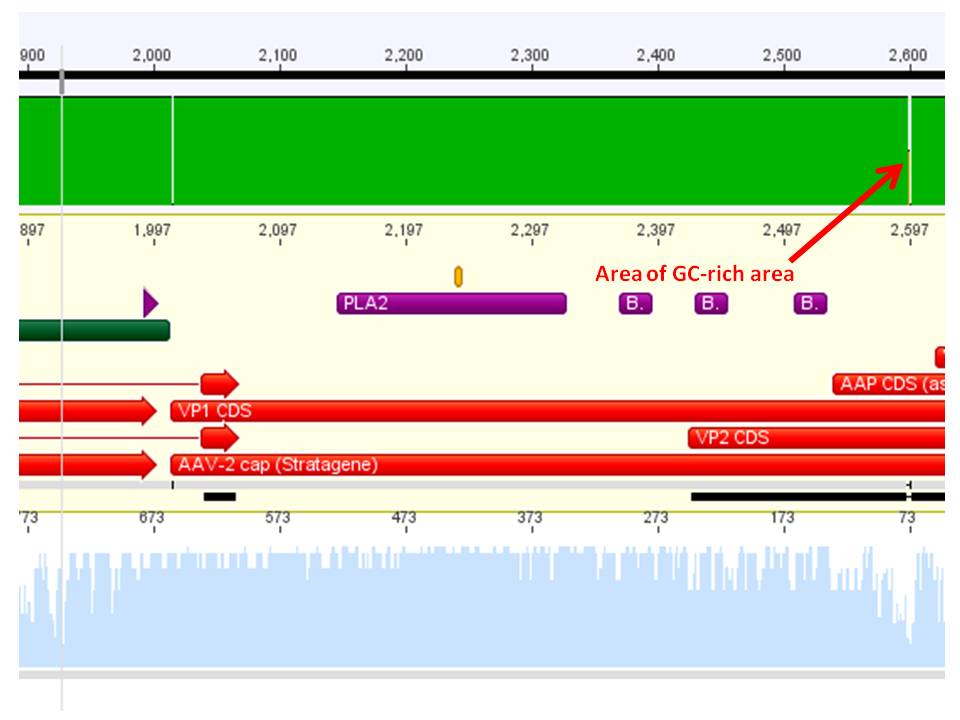
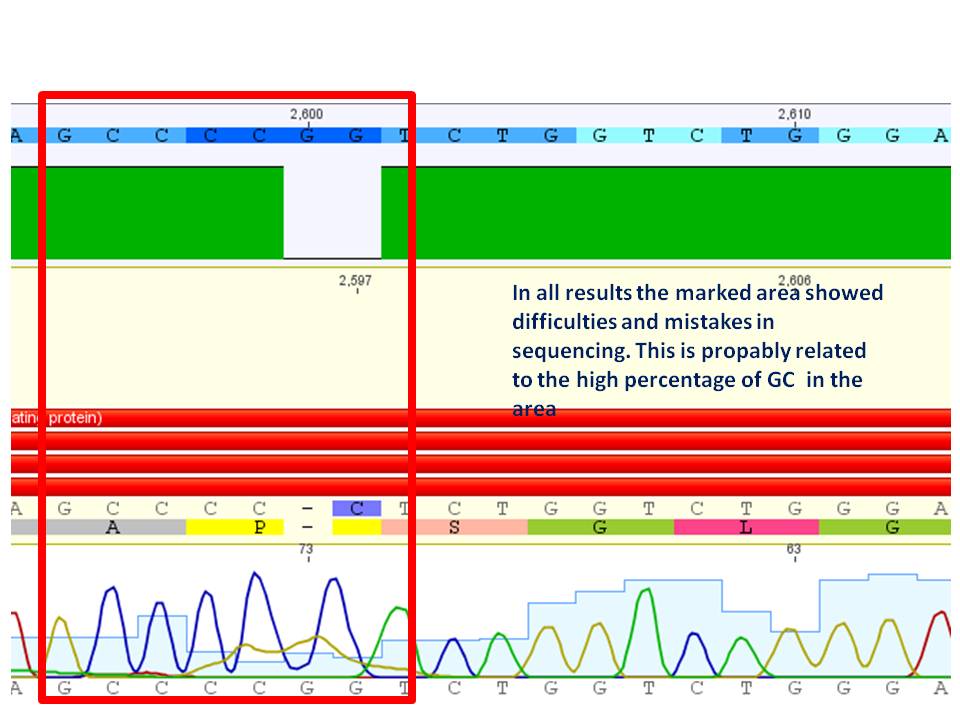
The synthetised Cap was inserted into the pAAV vector already containing the 4 mutations and the Rep construct.
- Tube name: SB1
- Primer used: VP 4200 rev
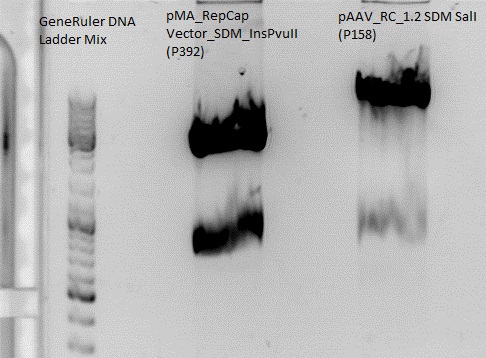
Cloning of Cap into pAAV_RC_1.2 SDM SalI
Investigator: Stefan
Comment: pAAV with inserted Rep does not work in cell culture as expected. It was assumed that this is related to the deletion of a restriction site for KpnI at the splicing site of Rep40/Rep68. To test our synthethised Cap a plasmid only containing this will be produced.
Digestion:
| components | pMA_RepCap Vector_SDM_InsPvuII (P392) | pAAV_RC_1.2 SDM SalI (P158) |
| DNA | 11,6 | 3,7 |
| Buffer 2 (10x) | 2 | 2 |
| XcmI | 1 | 1 |
| BsiWI | 1 | 1 |
| H2O | 4,4 | 12,3 |
| Total volume | 20 | 20 |
Comment:Digestion was performed using two steps, first incubating for 1 hour at 37 °C, afterwards for 1,5 hours at 55 °C.
Gel:
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 115 Volt, running time: 50 minutes
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- pMA_RepCap Vector_SDM_InsPvuII (P392): c= 8,43 ng/µl
- pAAV_RC_1.2 SDM SalI (P158) c= 17,24 ng/µl
T4 Ligation:
Comment: XcmI produces only 1 base overhang, but using Quick Ligase is not working properly. Therefore ligation is performed using T4.
The Ligation was performed as following:
- Vector Volume: 4,17 µl
- Insert Volume: 3,83 µl
- 1 µl T4 ligase buffer (10x)
- 8 µl (Vector + Insert) mix
- 1 µl T4 Ligase
Incubating for 70 minutes.
Transformation:
Trafo was performed according to the standard protocol (BL21). The cells were plated on a agar plate with ampicilin.
Repetition of Mini-Preps and test digestion of Linker BioBricks from 2nd ligation
Investigator: Achim, Anna
Comment: Recent update of succesfully produced ViralBricks:
File:Freiburg10 Production of ViralBricks.pdf
Test digestion:
| Number | 11.4 | 11.4 | 12.3 | 12.4 | 13.3 | 13.4 | 14.3 | 14.4 |
| DNA | 7 | 6,5 | 5,5 | 5 | 5 | 12,52 | 8,5 | 7 |
| H2O | 5,5 | 6 | 7 | 7,5 | 7,5 | 0 | 4 | 5,5 |
BSA: no
Enzymes:
EcoR I HF: 0,5 µl
Pst I HF: 0,5 µl
Total volume /µl:10
- Incubation time: 3/4 h, Incubation temperature: 37°
Preparation of gel:
- 1 g Agarose, 50 ml TAE (2%), 6 µl GELRED , at 115 Volt, running time: 45 minutes
To do: A new approach of the ViralBricks production has to be done tomorrow. A possible reason for the failure of the reactions is that the restriction enzymes didn't cut in the kenow buffer.
Preparation for Midi-Preps of pAAV_mVenus, pHelper and pAAV_RC
Investigator: Kerstin
- 25 ml DYT was prepared with 25 µl Ampicillin and inoculated with glycerol stocks of pAAV_mVenus, pHelper and RC, each were incubated over night in 37°C room.
Sequencing
Investigator: Hanna
pCerulean_Zegfr:1907_ShortLinker, pCerulean_Zegfr:1907_MiddleLinker, pCerulean_Zegfr:1907_LongLinker, pCerulean_Zegfr:1907_SEG and pCerulean_CFP_MiddleLinker were sent for sequencing.
Used primer: GATC_std_CMV-F
114. labday 08.09.2010
Sequencing results of pCerulean_Zegfr:1907_"Linker" and pCerulean_CFP_MiddleLinker
Investigator: Hanna
Comment: In order to perform single virus tracing experiments, CFP, which was fused to the Middle-Linker, was cloned into pCerulean. Referring to the establishment of a small linker library, the Affibody Zegfr:1907 was fused to the Short-Linker, Middle-Linker, Long-Linker and SEG-Linker and was then cloned into pCerulean.
Sequencing results:
1. pCerulean_CFP_MiddleLinker:
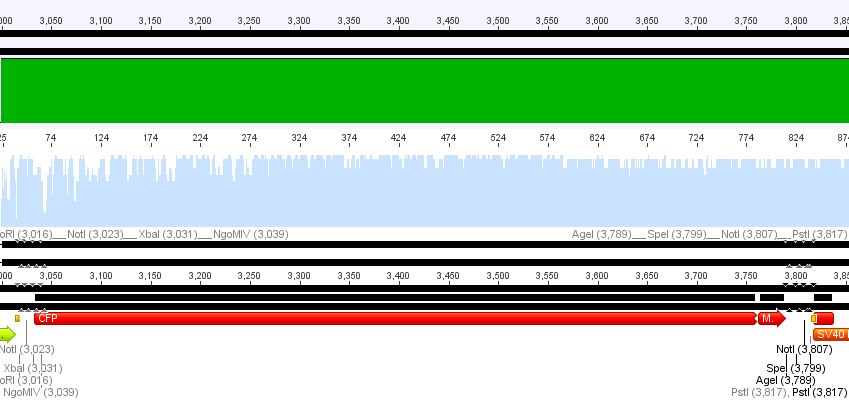
Conclusion: Sequencing looked well, CFP_MiddleLinker was cloned successfully into pCerulean is now ready for fusion to VP2/3.
2. pCerulean_Zegfr:1907_ShortLinker:
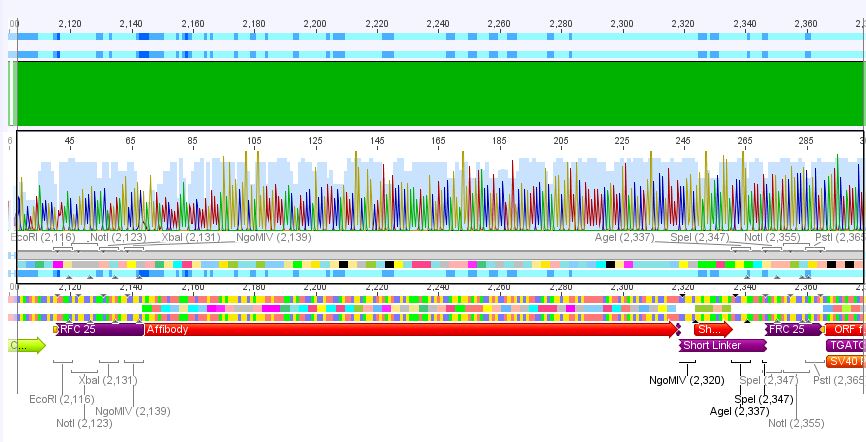
Conclusion: Sequencing looked also well, Zegfr:1907_ShortLinker was cloned successfully into pCerulean is now ready for fusion to VP2/3.
3. pCerulean_Zegfr:1907_MiddleLinker:
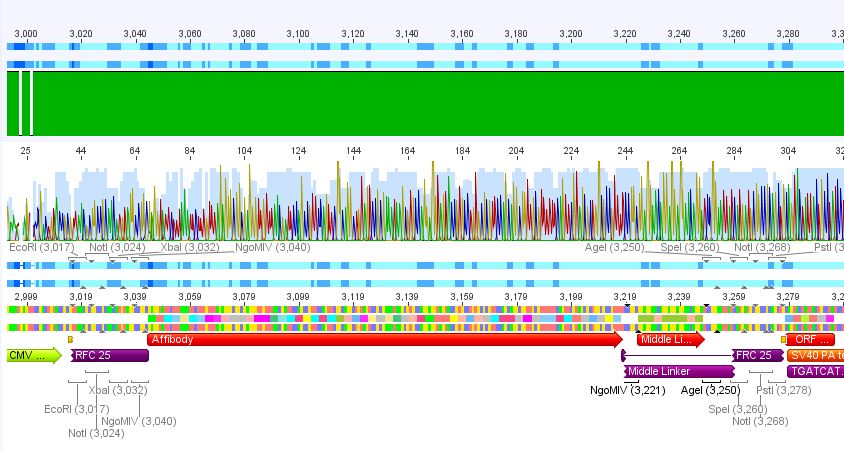
Conclusion: Sequencing looked also well, Zegfr:1907_MiddleLinker was cloned successfully into pCerulean is now ready for fusion to VP2/3.
4. pCerulean_Zegfr:1907_LongLinker:
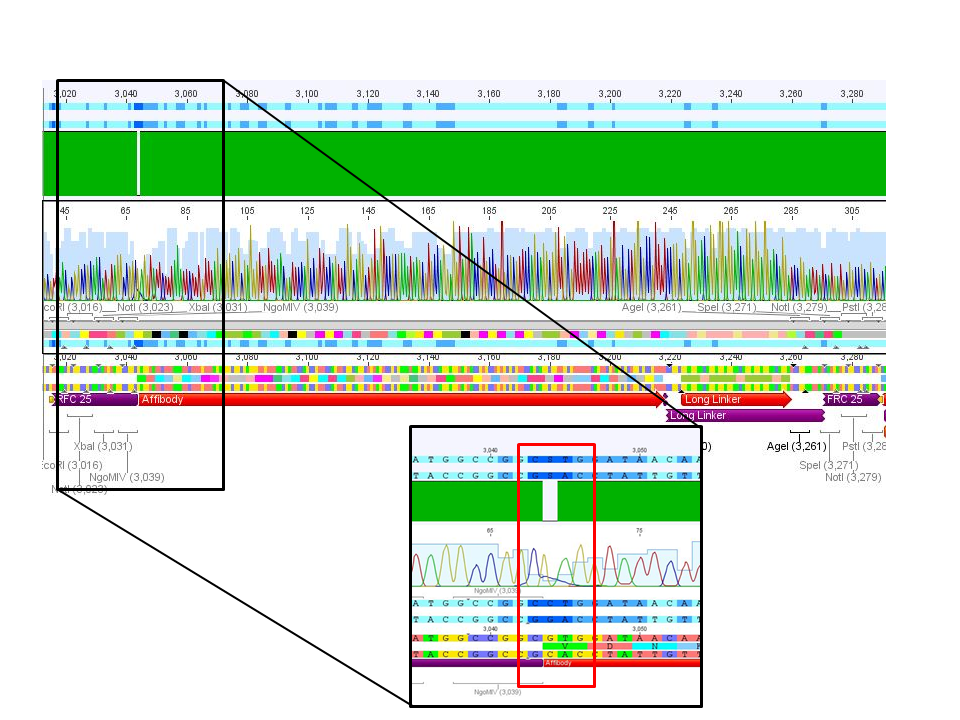
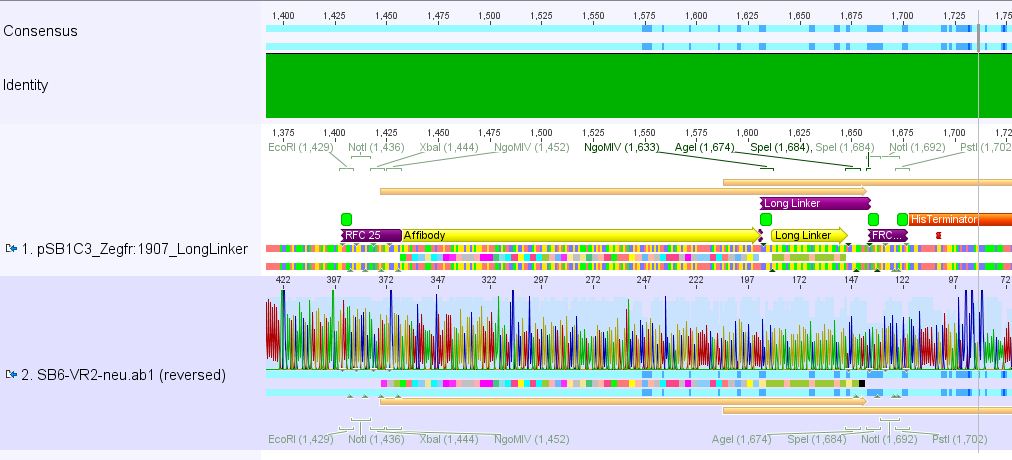
Conclusion: Sequencing delivered that there was a C -> G transistion between the NgoMIV restriction site and the Zegfr:1907 sequence. Because it's located in the gene-synthesis region, the question is, whether there was a mutation caused by the XL1b cells or whether sequencing quality is not good enough. Therefore it was decided to sequence this conbstruct once again in reverse direction.
Conclusion: Sequencing looked also well, Zegfr:1907_SEG was cloned successfully into pCerulean is now ready for fusion to VP2/3.
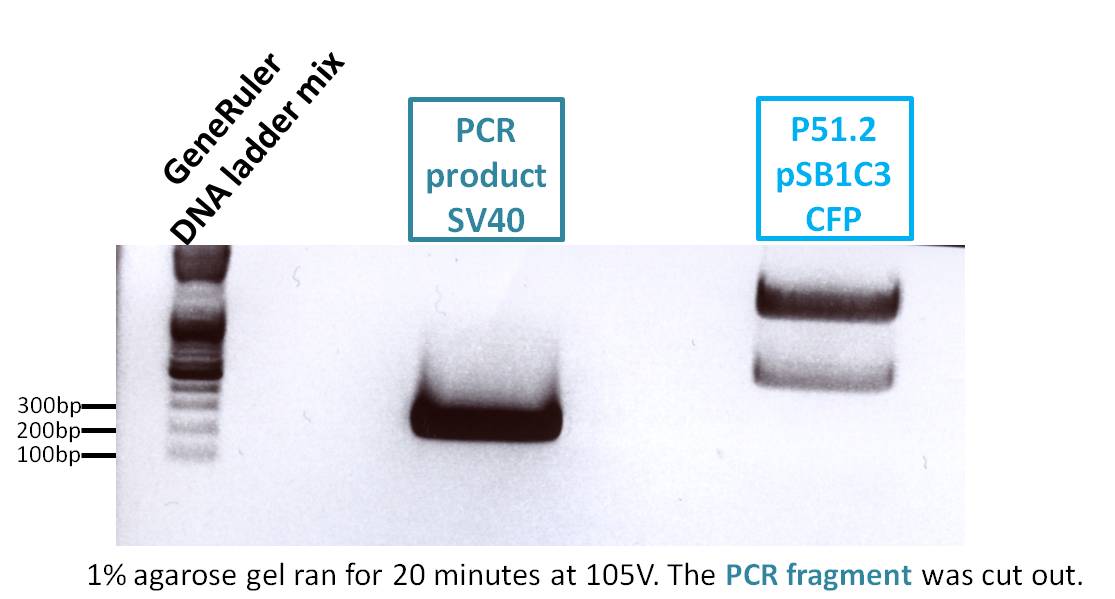
Repetition of PCR of SV40 terminator
Investigator: Anna
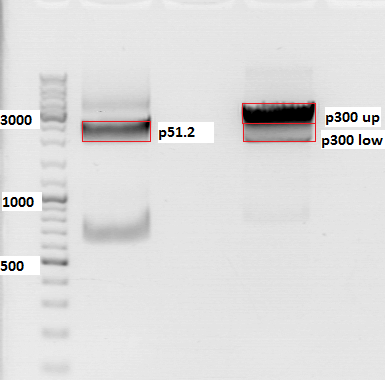
Comment: Second repetition of SV40 PCR, insert and vector were cut with Age I and Xba I.
PCR sample: p230 (PeGFP_C1)
Plasmid backbone: p51.2 (pSB1C3_CFP)
| Ingredients | CD sample |
| 5X Phusion HF buffer | 10 µl |
| 10 mM dNTP mix | 1µl |
| forward primer: O158 | 2,5 µl |
| reverse primer: O159 | 2,5 µl |
| DNA Template | 3 µl |
| DMSO | 0 µl |
| Phusion Polymerase | 0,5 µl |
| H2O | 30,5 µl |
| Total volume | 50 µl |
PCR program:
| Cycles | Temperature | Time |
| 98°C | 1 | |
| 8x | 98°C | 15" |
| 59°C | 25" | |
| 72°C | 7" | |
| 17x | 98°C | 15" |
| 67°C | 25" | |
| 72°C | 7" | |
| 1x | 72°C | 5' |
| Hold 4°C |
Digestion of plasmid backbone:
c (pSB1C3_CFP) = 151, 1 ng/ µl
| Components | vector Volume/µL |
| DNA 1 µg | 7,0 µl |
| BSA (100x) | 2,0 µl |
| Buffer no. 4 (10x) | 2,0 µl |
| Enzyme 1 XbaI | 0,5 µl |
| Enzyme 2 AgeI HF | 0,5 µl |
| H2O | 8 µl |
| Total volume | 20 |
incubation @ 37 °C for approx. 2,5 h
Preparation of gel:
0,75 g Agarose, 50 ml TAE (1,5%), 6 µl GELRED , at 115 Volt, running time: 45 minutes
Expected size of fragment: 261 bp
Digestion of PCR product:
| Components | PCR product Volume/µL |
| DNA | 18,5 µl |
| BSA (100x) | 3 µl |
| Buffer no. 4 (10x) | 3 µl |
| Enzyme 1 XbaI | 1 µl |
| Enzyme 2 AgeI HF | 1 µl |
| H2O | 3,5 µl |
| Total volume | 30 |
incubation @ 37 °C for approx. 2 h
Ligation
c(pSB1C3_cut_Age_Xba) = 0,9 ng/µl
c(Insert) = 25,8 ng/µl
T4 Ligase was used.
DNA-mix 8 ul (7,08 µl insert + 0,92 µl insert)
T4 Ligase 1µl
T4 Ligase Buffer 1 µl
Incubation @ RT for 20'
Transformation
Transformation was done according to the standard protocol with XL1B cells.
PCR of P40
Investigator: Stefan
DNA sample was diluted 1:1000
PCR progam:
| Ingredients | Volume / µl |
| 5X Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| forward primer: O152 | 2,5 |
| reverse primer: O153 | 2,5 |
| DNA Template | 1 |
| DMSO | - |
| Phusion Polymerase | 0,5 |
| H2O | 31,5 |
| Total volume | 50 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 98 | 60 | |
| 8x | 98 | 15 |
| 61 | 25 | |
| 72 | 3 | |
| 17x | 98 | 15 |
| 69 | 25 | |
| 72 | 4 | |
| 1x | 72 | 300 |
| Hold | 4 |
Digestion of plasmid backbone:
Plasmid used: pSB1C3_CFP (P51.2)
c (pSB1C3_CFP) = 151, 1 ng/ µl
| Components | vector Volume/µL |
| DNA | 10 |
| BSA (10x) | 2 |
| Buffer no. 4 (10x) | 2 |
| XbaI | 1 |
| PstI | 4 |
| H2O | 4 |
| Total volume | 20 |
incubation @ 37 C for approx. 2 h
Comment: PCR does not work out twice. Therefore troubleshouting will have to be done.
Midi-Prep
Investigator: Anissa, Kerstin
Midi-Prep was performed with B34 (pAAV_iGEM_mVenus_YFP), R/C (standard P357) and pHelper (standard P356) according to the standard protocol.
- c(pHelper): 518,7 ng/µl
- c(RC): 1068,7 ng/µl
- c(mVenus): 277,9 ng/µl
Seeding HT1080, Passaging AAV293 and Transfection
Investigator: Kerstin, Patrick
- Seeding HT1080 cells for Transduction at 09.09.2010: two 6-well plates were prepared (200.000 cells per well)
- Passaging AAV293 cells from T75 to T175 flasks
- Transfection of AAV293: Ten plates same treatment (10µg of each plasmid):
- c(RC)= 1068,7 ng/µl --> 9,4 µl
- c(pHelper)= 518,7 ng/µl --> 19,3 µl
- c(TKGMK, P81b)= 1900 ng/µl --> 5,3 µl
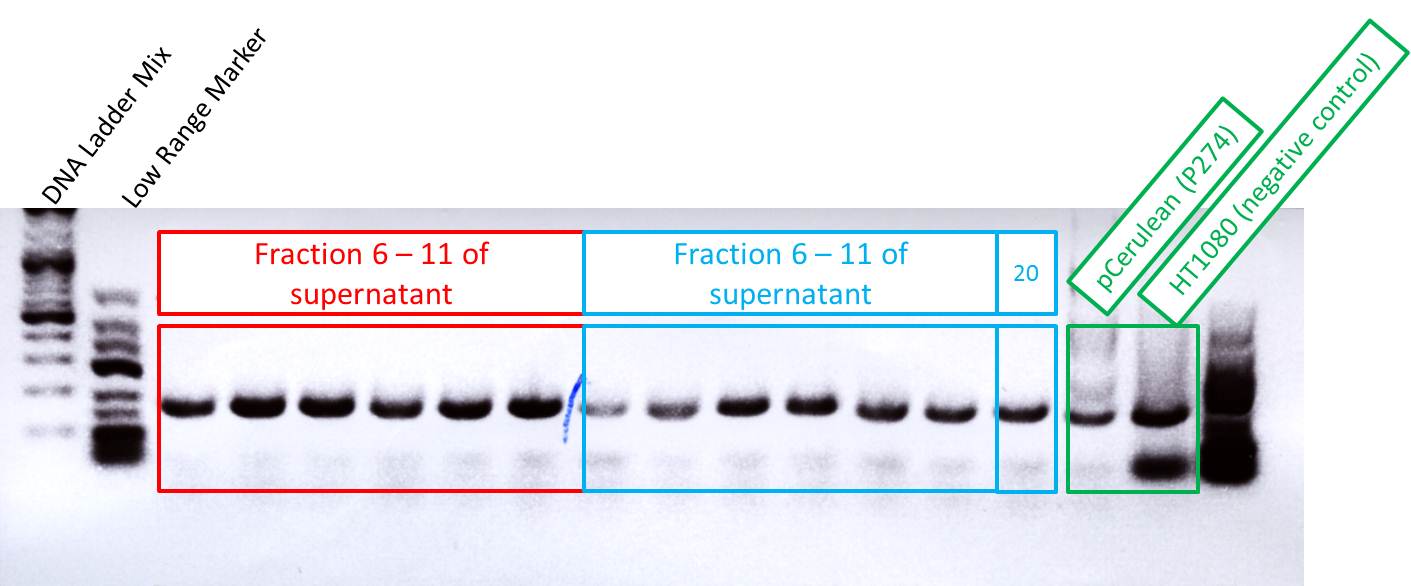
Checking fractions of Äkta run
Investigator: Stefan
Comment: Fractions 6 - 11 of supernatant and of lysated cells treated with benzonase have to be checked for virus particles as well as fraction 20 of the second approach. This will be done amplifing the CMV promotor. Additionally, one positive control (pCerulean P274) and one negative control (HT1080 cells already confirmed being negative in qPCR) were used for PCR approach.
PCR progam:
primers used:
| Ingredients | Volume / µl |
| 10x ThermoPol buffer | 10 |
| 10 mM dNTP mix | 1 |
| forward primer: O20 | 2,5 |
| reverse primer: O21 | 2,5 |
| MgCl2 (50mM) | 4 |
| fraction | 5 |
| DeepVent | 0,5 |
| H2O | 8 |
| Total volume | 25 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 95 | 600 | |
| 35x | 95 | 20 |
| 56 | 20 | |
| 72 | 18 | |
| 1x | 72 | 120 |
| Hold | 4 |
Gel run will have to be performed tomorrow.
115. labday 09.09.2010
Picking clones of leftITR_hTERT_beta-globin and leftITR_CMV_beta-globin biobricks
Kira
The plates were checked in the morning and they did not contain any visible colonies, thus the plates were kept @ 37C till the late afternoon in order to check again.
Fortunately, after approx. 18h of incubation the plate observation revealed lots of colonies, thus 3 clones from each plate could be inoculated in the DYT media.
new ligation try and transformation of CD-biobrick
Kira
CD-biobrick plate contained just few (around 3 colonies), thus new ligation approach was performed. T4 ligase was used. The ligation mix was incubated @ 18C for around 5 hours
Transformation was performed according to the standard protocol
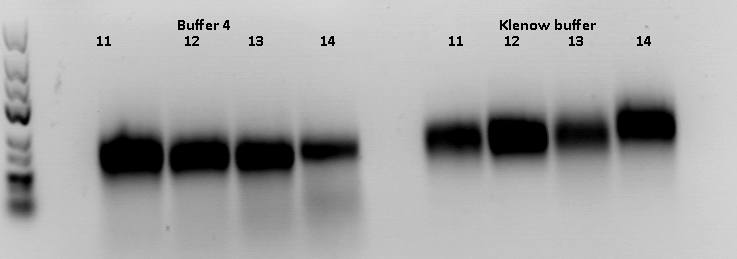
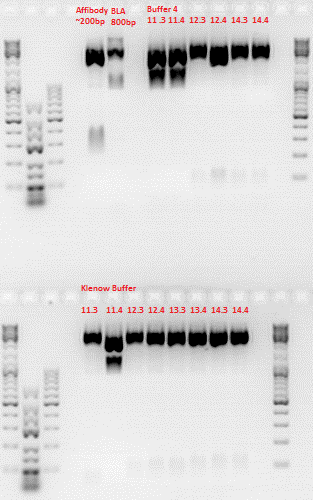
Repetition of ViralBrick Ligations: 453 Z34C, 587 Z34C, 587 KO Z34C, 587 KO Z34C Spacer
Investigator: Achim
- Nomenclature:
11K: 453_Z34C in Klenow Buffer
12K: 587_Z34C in Klenow Buffer
13K: 587_KO_Z34C in Klenow Buffer
14K: 587_KO_Z34C_Spacer in Klenow Buffer
11 4: 453_Z34C in Buffer 4
12 4: 587_Z34C in Buffer 4
13 4: 587_KO_Z34C in Buffer 4
14 4: 587_KO_Z34C_Spacer in Buffer 4
Two new reaction setups for the remaining four ViralBricks:
- Hybridisation & Fill-In reaction in Buffer 4, heat inactivation, digestion, ligation
- Hybridisation & Fill-In reaction in Klenow Buffer, heat inactivation, PCR Purification, digestion, ligation
We followed the recommendations provided by Fermentas with their Klenow Fragment manual:
- 2 µl total DNA (3,3 µl per Oligo)
- 0,1 µl dNTPs
- 2 µl Klenow Buffer/ Buffer 4
in a 20 µl reaction.
| Components | 453 Z34C | 587 Z34C | 587 KO Z34C | 587 KO Z34C SPACER |
| O143 | O145 | O147 | O149 | |
| O144 | O146 | O148 | O150 |
Hybridisation was performed using the TKfill program on the gradient cycler. After Hybridisation, 0,5 µl Klenow fragment was added. The fill-in reaction was run for 40 minutes at 37°C. Afterwards, the Klenow fragment was heat inactivated at 75°C for 10 minutes. The samples containing the Klenow buffer were then purified with the QIAquick PCR Purification Kit Protocol.
Samples 11 K and 11 4 were digested with SspI and SalI, Samples 12-14 were digested with BamHI and PvuII. Buffer 4 was added to the purified samples. One sample of the vector plasmid pSB1C3_BLA (P320) was digested with SspI and SalI, two samples were digested with BamHI and PvuII two obtain a sufficient amount of DNA for 6 ligations.
(Table with concentrations and used DNA Volumes)
After digestion, the digested inserts were separated on a 2% agarose gel, the vector backbones were separated on a 1% agarose gel:
(Pictures of gels)
(Nanodrop concentrations of gelex:)
Ligation:
(Ligation volumes)
Vectors and Inserts were ligated for ~3h with T4 Ligase at room temperature.
Two µl of each ligation were transformed into XL1B cells according to our standard protocol. Plates were incubated overnight
Midiprep of pHelper, pAAV_RC and pAAV_iGEM_mVenus (B34)
Investigator: Patrick
The Midiprep was performed according to the standard protocol yieldung the following concentrations:
pAAV_iGEM_mVenus (B34): 1131,7 ng/µl, labeled P419,420,421
pHelper: 548 ng/µl
pAAV_RC: 985 ng/µl
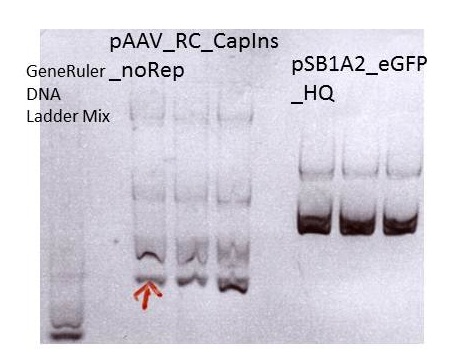
Miniprep of pAAV_RC_CapIns_noRep and pSB1A2_EGFP_HQ
Investigator: Stefan
Mini-Prep was performed according to the standard protocol
| components | pAAV_RC_CapIns_noRep clone1-3 | pSB1A2_EGFP_HQ clone1-3 |
| DNA | 1 | 1 |
| Buffer (10x) | 1 | 1 |
| BSA (10x) | 1 | 1 |
| Enzyme 1 | Acc65I 0,5 | XbaI 0,5 |
| Enzyme 2 | BsiWI 0,5 | PstI 0,5 |
| H2O | 6 | 6 |
| Total volume | 10 | 10 |
Sent for sequencing: pAAV_RC_CapIns_noRep clone1 (P413)
- Tube name: SB1
- Primer used: VP 4200 rev
Sent for sequencing: pSB1A2_EGFP_HQ clone1 (P416)
- Tube name: SB2
- Primer used: VR2
PCR of VP2/VP3

Comment: VP2/3 will be separated from cap for n-terminal fusion
Investigator: Jessica
DNA sample was diluted 1:1000
- P357 = pAAV_RC
- P396 = pAAV_RC_CapIns_prepSDM clone 3
PCR progam:
| Ingredients | Volume / µl | Volume / µl |
| 5X Phusion HF buffer | 10 | 10 |
| 10 mM dNTP mix | 1 | 1 |
| forward primer: O90 | 2,5 | 2,5 |
| reverse primer: O120 | 2,5 | 2,5 |
| DNA Template | 0,78 | 2 |
| DMSO | 1 | 1 |
| Phusion Polymerase | 0,5 | 0,5 |
| H2O | 31,72 | 30,5 |
| Total volume | 50 | 50 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 98 | 60 | |
| 8x | 98 | 15 |
| 61 | 25 | |
| 72 | 30 | |
| 17x | 98 | 15 |
| 72 | 25 | |
| 72 | 30 | |
| 1x | 72 | 300 |
| Hold | 4 |
Digestion of plasmid backbone:
Plasmid used: pSB1C3_RFC_SEG (P107)
c (pSB1C3_RFC_SEG) = 185,6 ng/ µl
| Components | vector Volume/µL |
| DNA | 5,4 |
| BSA (10x) | 2 |
| Buffer no. 4 (10x) | 2 |
| NgoMIV | 1 |
| SpeI | 1 |
| H2O | 9,6 |
| Total volume | 20 |
incubation @ 37 C for approx. 2h
1% agarose gel
Digestion of PCR product:
| Components | PCR product Volume/µL |
| DNA | 30 |
| BSA (10x) | 4 |
| Buffer no. 4 (10x) | 4 |
| NgoMIV | 1 |
| SpeI | 1 |
| H2O | - |
| Total volume | 40 |
incubation @ 37 C for approx. 2,5h
Ligation:
1 µl T4 DNA ligase, 1 µl 10x Buffer, µl vector (P107), µl Insert (P357)/ µl Insert (P396).
Incubation time: over night
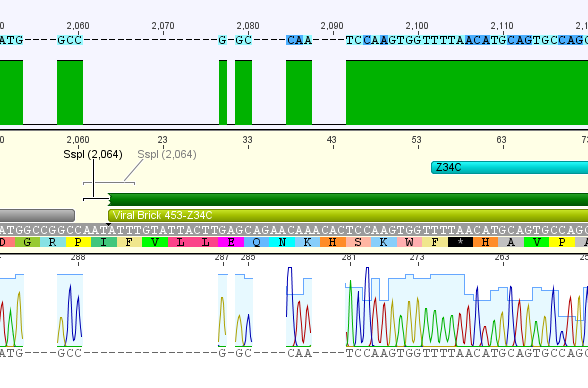
Sequencing results of pCerulean_VP1up_NLS_Affibody/mVenus/Histag
Investigator: Anna
Comment: All results were classified in agreement with Sven. They can be used for generating the full construct (with VPex3).
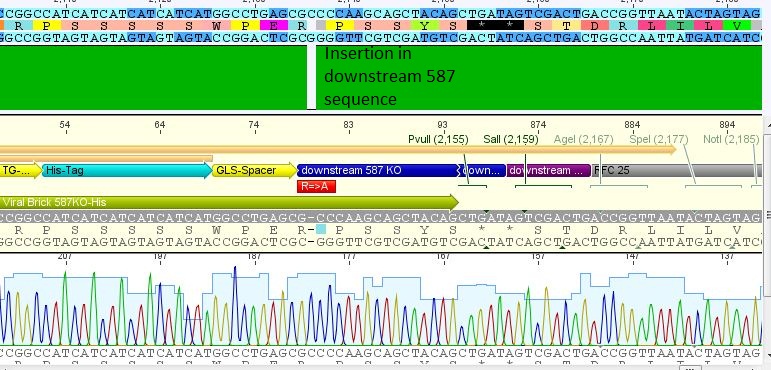
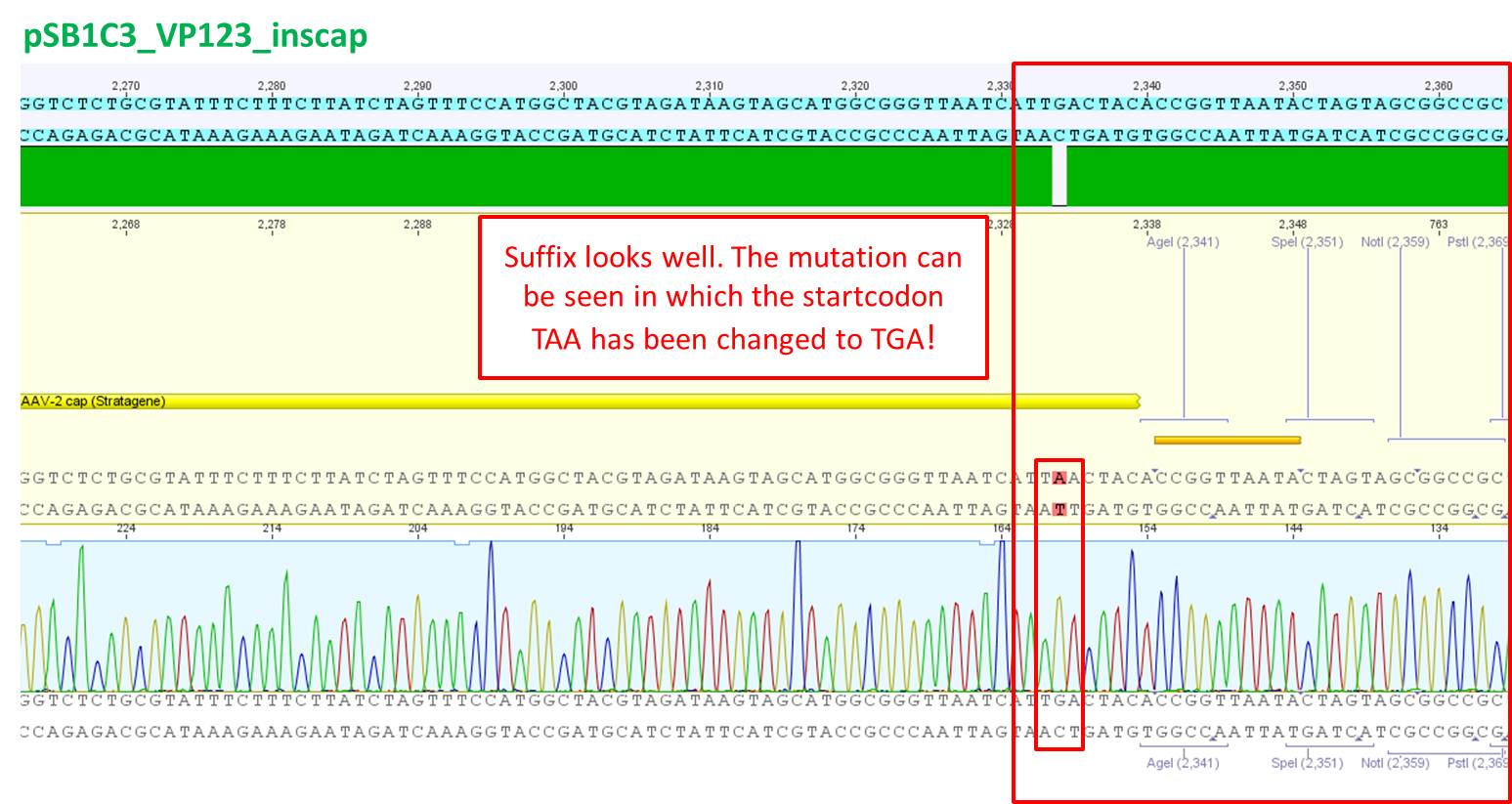
pCerulean_VP1up_NLS_Affibody: Insert ok, but there is a G -> C transition.
pCerulean_VP1up_NLS_mVenus: The NgoMIV restriction site is present, but as above, there is a G -> C transition. Third mutation downstream is a sequencing fault, there are clearly two peaks for C.
pCerulean_VP1up_NLS_Histag: Insert was succesfully cloned into p_Cerulean.
Biobrick production of leftITR-phTERT-beta-globin and leftITR_pCMV-beta-globin
Motivation: Production of fusion proteins
Investigator: Kira
Digestions of backbones and inserts:
c (pSB1C3_leftITR_phTERT) = 179 ng/ µl
c(pSB1C3_leftITR_pCMV) = 231 ng/ul
| Components | TERT | CMV | beta-globin |
| DNA | 5,5 | 4,5 | 8,5 |
| BSA (100x) | 0,25 | 0,25 | 0,25 |
| Buffer no. 4 (10x) | 2,5 | 2,5 | 2,5 |
| PstI-HF | 0,5 | 0,5 | 0,5 |
| SpeI | 1 | 1 | 1 |
| XbaI | 0 | 0 | 1 |
| H2O | 15,25 | 16,25 | 12,25 |
| Total volume | 25 | 25 | 25 |
incubation @ 37 C for approx. h
1% agarose gel
Ligation:
Quickligase was used
TERT-mix 9 ul (5,3 ul vector + 3,7 ul insert)
CMV-mix 9 ul (3,7 ul vector + 5,3 ul insert)
Quickligase 1ul
Quickligase Buffer 10 ul
Incubation @ RT for 10'
Transformation:
Transformation was performed according to the standard protocol w XL1B cells (thanks Achim!)
Transfection, Harvest viral particles and Transduction
Investigator: Kerstin
- Transfection of 15x 6-well plates (each plate same treatment --> 10 µg each plasmid)
- Harvest viral particles (from 06.09.2010) and Transduction of HT1080 cells
1.plate:
- 1.1 no virus
- 1.2 lITR_CMV_beta-globin_mVenus_rITR
- 1.3 lITR_CMV_beta-globin_mVenus_rITR
- 1.4 lITR_CMV_beta-globin_mVenus_rITR
- 1.5 lITR_CMV_beta-globin_mVenus_Hgh_rITR
- 1.6 lITR_CMV_beta-globin_mVenus_Hgh_rITR
2.plate:
- 1.1 no virus
- 1.2 lITR_CMV_mVenus_Hgh_rITR
- 1.3 lITR_CMV_mVenus_Hgh_rITR
- 1.4 lITR_CMV_mVenus_Hgh_rITR
- 1.5 lITR_CMV_beta-globin_mVenus_Hgh_rITR
- 1.6 lITR_CMV_beta-globin_mVenus_Hgh_rITR
Competent XL1-B cells
Investigator: Patrick
Just for you! ;-)Do you know the normal transformation efficiency of chemically competent cells? What is good, waht is bad?? Thanks!
Today i checked the mutual competent cells i made for their transformation ability. It is ok (>10^6 cells / µg DNA).
Now there are about 100 new aliquots (each 100 µl) ready to use.
Checking PCR amplified virus particles
Investigator: Bea
Comment: The virus particles purifed by using liquid chromatography ("ÄKTA")(containing which construct??) were PCR- amplified (dame primers were used as for the qPCR which were already tested and could be confirmed for proper working) in order to obtain insights into which collected fraction does contain our virus particels. Therefore the PCR amplified samples were loaded on an 2% agarose gel.
- Virus particle containing following plasmid: pAAV_iGEM_mVenus
- Liquid chromatography performed by: Volker
- PCR amplification performed by: Stefan
Results:
As it can be seen in the picture, the expected PCR-fragment size of 200 bp can be detected in each fraction. Fractions collected 6 to 11 of the supernatant after cell lysis and centrifugation and fractions collected 6 to 11 of the cell pellet after cell lysis and centrifugation. The fraction 20 of the cell pellet was aswell PCR amplified and loaded on the agarose gel. The postivie control (added the plasmids containing a CMV promoter which were sequenced and confirmed) is loaded (pCerulean P274) and the negative control which contained only harvested HT1080 cells (harvested, but whithout any transduction) can be seen in lane 16 and 17, respectively. Since the PCR has been performed with a low TM and 35 cycles this could be the reason for the detection of the CMV promoter of the negative control HT1080. next time PCR should be carried out with only 25 cycles and a higher TM= 60°C. But the detection of the CMV promoter by using a normal PCR does work.
Quickchange site directed mutagenesis of pAAV_RC_CapIns_prepSDM
Investigator: Stefan
Comment: In cellculture, pAAV with inserted synthesised Rep did not work out. It was suggested that this could be related to an insertion of a KpnI restriction site near a splicing site. To verify this, the restriction site needs to be removed and tested in cellculture again.
Ingredients:
| Ingredients | Volume / µl |
| 10x reaction buffer | 2,5 |
| dNTP | 0,5 |
| forward primer: O182 | 0,52 |
| reverse primer: O183 | 0,52 |
| DNA Template | 2 |
| QuikSolution Reagent | 0,75 |
| QuikChangeLightning Enzyme | 0,5 |
| H2O | 17,71 |
| Total volume | 25 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 95 | 120 | |
| 8x (step 2-4 | 95 | 15 |
| 60 | 10 | |
| 68 | 225 | |
| 68 | 600 |
After PCR, the product was incubated with DpnI at 37 °C for 10 minutes.
Transformation:
Transformation was performed according to standard protocol using XL10-Gold cells. Cells were plated on plate containing Ampicilin.
116. labday 10.09.2010
Trafo evaluation of CD-biobrick plate
Investigator: Kira
Compared to the last tries, the ligation with T4 ligase was much more successful. The plate containes colonies.
Picking clones of yesterdays ViralBrick ligations and of Jessicas ligation
Investigator: Achim
Seeding HT1080 cells
Investigator: Kerstin
- Ten 6-well plates of HT1080 cells were seeded (Transduction at 11.09.2010)for testing the optimal concentration of ganciclovir
Test digestion of pSB1C3_SV40_Terminator_Clone1-3
Investigator: Anna
| Samples | Insert | Clone | Concentration [ng/µl] |
| 1 | pSB1C3_SV40_Terminator | 1 | 204,2 |
| 2 | pSB1C3_SV40_Terminator | 2 | 246,1 |
| 3 | pSB1C3_SV40_Terminator | 3 | 200,9 |
Test digestion:
| components | Volume for each sample /µl |
| DNA | 3 |
| BSA (10x) | 1 |
| Buffer 4 (10x) | 1 |
| Enzyme HF | 0,5 |
| Enzyme HF | 0,5 |
| H2O | 4 |
| Total volume /µl | 10 |
- Incubation time: 1,5 h, Incubation temperature: 37°
Preparation of gel:
- 0,5 g Agarose, 50 ml TAE (1%), 6 µl GELRED , at 115 Volt, running time: 40 minutes
Comment: As shown on the gel, the vector backbone was religated with CFP (size: ~730 bp). PCR and Cloning steps have to be repeated, because the wrong restrition enzymes were used.
Sequencing analysis of pSB1A2_eGFP_HQ and pAAV_RC_CapIns_noRep
Investigator: Stefan
pSB1A2_eGFP_HQ
Comment: To improve iGEMs eGFP, it will be converted into RFC25 Freiburg standard. Plasmid preped from retrafo was sent for sequencing to verify sequence.
pAAV_RC_CapIns_noRep
Comment: Since pAAV containing the synthetised Rep does not work out in cell culture an additional approach was performed inserting only the synthesized Cap and test this.
Mini-prep and test digestion of leftITR_hTERT_beta-globin and leftITR_CMV_beta-globin biobricks
Investigator: Kira
Mini-Prep was performed with 3 clones of each construct in order to continue with test digestion and sequencing.
Test digestion:
| components | Volume for each sample /µl |
| DNA | 2.5 |
| BSA (10x) | 2 |
| Buffer 4 (10x) | 2 |
| Enzyme Xba | 1 |
| Enzyme PstI-HF | 0,5 |
| H2O | 12 |
| Total volume /µl | 20 |
1% agarose gel
CMV_clone1 as well as hTERT_clone1 were sent for sequencing
117. labday 11.09.2010
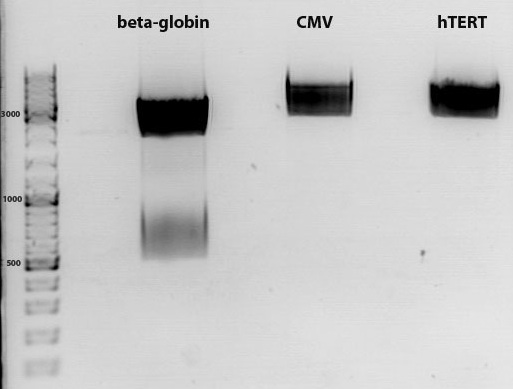
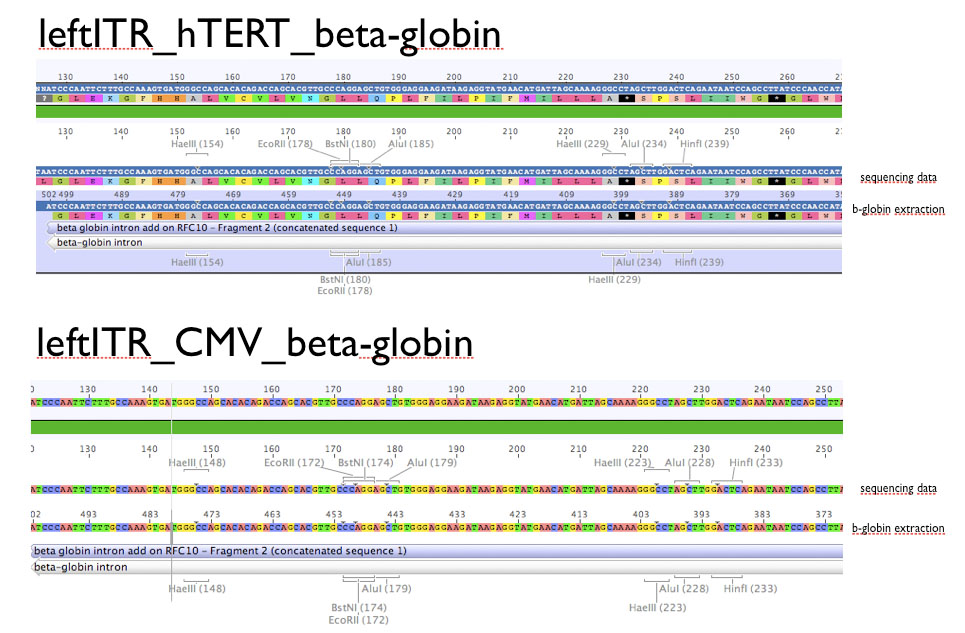
Sequencing evaluation of leftITR_CMV_beta-gloin and leftITR_hTERT_beta-globin biobricks
Investigator: Kira
The sequencing reveals that both biobricks were seccessful cloned.
Comment: For the evaluation, beta-globin intron was extracted from the original beta-globin biobrick and checked against the sequencing data (no connection to the lab server at home. the 'single' biobricks are only data, which are saved on my mac)
FACS analysis, harvest viral particles and Transduction
Investigator: Kerstin
- FACS analysis of Constructs with/without Hgh/ß-globin
- Harvested viral particles of transfection from 08.09.2010
- Transduction for testing the optimal concentration of ganciclovir
Mini-Preps of Z34C ViralBricks
Investigator: Anna
Recent update of ViralBricks: The hybridisation and fill-in reactions of the Z34C constructs were repeated with heat inactivation of the Klenow enzyme (see labday 04.09.10), without usable results. In addition, a second ligation of 453_Bap was done with succes. This time two new approaches of the fill-ins were prepared (labday 09.09.10), one by using the Klenow Buffer and one with the Standard NEB Buffer 4.
Buffer 4:
| Samples | Insert | Clone | Concentration [ng/µl] |
| 11.4.1 | 453_ Z34C | 1 | 271,67 |
| 11.4.2 | 453_ Z34C | 2 | 218,01 |
| 12.4.1 | 587_Z34C | 1 | 199,88 |
| 12.4.2 | 587_Z34C | 2 | 357,13 |
| - | 587_KO_Z34C | - | - |
| - | 587_KO_Z34C | - | - |
| 14.4.1 | 587_KO_Z34C_Spacer | 1 | 172,76 |
| 14.4.2 | 587_KO_Z34C_Spacer | 2 | 337,54 |
Klenow Buffer:
| Samples | Insert | Clone | Concentration [ng/µl] |
| 11.1 K | 453_ Z34C | 1 | 220,89 |
| 11.2 K | 453_ Z34C | 2 | 176,34 |
| 12.1 K | 587_Z34C | 1 | 198,73 |
| 12.2 K | 587_Z34C | 2 | 182,64 |
| 13.1 K | 587_KO_Z34C | 1 | 186,50 |
| 13.2 K | 587_KO_Z34C | 2 | 158,51 |
| 14.1 K | 587_KO_Z34C_Spacer | 1 | 182,25 |
| 14.2 K | 587_KO_Z34C_Spacer | 2 | 180,06 |
Mini-Preps of separated VP2/3 for N-terminal fusion
Investigator: Anna
- P357 = pAAV_RC
- P396 = pAAV_RC_CapIns_prepSDM clone 3
| Samples | Clone | Concentration [ng/µl] |
| 357.1 | 1 | 290,74 |
| 357.2 | 2 | 282,97 |
| 357.3 | 3 | 307,63 |
| 396.1 | 1 | 302,04 |
| 396.2 | 2 | 314,58 |
| 396.3 | 3 | 298,77 |
Update and next steps of daily labwork
Investigators: Bea, Stefan
Preparation of pAAV_RC construct
- We inserted the synthesized “rep” gene into the pAAV_RC construct with the four mutated restriction sites PstI (positions 310 and 4073), SalI (position 1239) and BamHI (position 856). The construct was tested in cell culture and no YFP expression of the vectorplasmid could be detected. Therefore, several new approaches were conducted.
- The first approach was to remutate the only “unwanted” restriction site(KpnI) in the supposed regulatory region of the rep gene. The basic idea was that we delete the KpnI site because KpnI was in the 453 loop in which we could insert additional small motifs. But, this idea was dismissed after some more research. Therefore the KpnI restriction site does not have to be deleted in the rep sequence. This construct will be tested in cell culture.
- Second, the experiment conducted in cell culture will be repeated with the same construct in order to exclude errors and mistakes related to the realization in cell culture.
- Since we cannot be sure if the inserted rep containing the remutated KpnI restriction site still works in cell culture, we decided to insert the synthesized “cap” into the already tested and confirmed pAAV_RC containing only the four mutated restriction sites (see above). This construct will be tested in cell culture aswell.
Overview:
| no plasmid number yet | P158 | P250 | P396 | P413 |
| pAAV_RC (4fold mutated, ins rep, SDM KpnI, inserted “cap”) | pAAV_RC (4-fold mutated) P158 | pAAV_RC (4fold. Inserted “rep”)P250 | pAAV_RC (4fold mutated, rep, cap)P396 | pAAV_RC ((4fold mutated, cap) P413 |
| not yet tested in cell culture | tested in cell culture and works | tested in cell culture, does not work. Experiment will be repeated | will not be tested in cell culture -> was used for SDM in rep gene. | has to be tested P413 |
| desired construct. Clones were picked today!! | contains many restriction enzymes. Is not the construct of choice | If second experiment fails again, will not be used then. | was only used for SDM in rep gene. | If cell culture experiments work --> synthesized cap still functions properly. |
N-terminal fusion
The constructs for the VP1 insertion and the Vp-2 fusion are ready for the next cloning step which means that we only need to fuse the VP2/3 PCR construct to the engineered plasmids containing different approaches. Two strategies will be followed until now:
- The first strategy is to PCR amplify the VP2/3 construct from the pAAV_RC (4fold mutated, P357 in order to fuse the working VP2/3 contruct to the targeting approaches.
was performed by Jessica (9.9.10), waiting for results
- The second strategy is to use the pAAV_RC (4fold mutated, ins rep, cap, P396) to PCR amplify the desired construct VP2/3.
was performed by Jessica (9.9.10), waiting for results
- The next step is to PCR amplify the VP2/3 aswell from the construct pAAV_RC (4fold mutated, ins rep, SDM KpnI, inserted “cap”)
plasmid should be ready tomorrow (12.9.10), can be performed afterwards
Comment (Hanna): We want to amplify VP2/3 from pAAV_RC (it's indifferent, whether the PCR is done with the "WT"-pAAV_RC or with the pAAV_RC- 4fold mutated... (the mutations are in rep!) Further on, we want to PCR amplify VP2/3 from pAAV_RC_capins and from pAAV_RC_capIns_HSPG-KO!!! Did you mean that?
Yes you are right! Thanks! We should keep that in mind, and do not forget about the HSPG knockout!!! (Bea) ;-)
Additionally, a site-directed mutagenesis has to be performed in order to obtain the plasmids which can be co-transfected with the modified construct containing the targeting molecules. Aim is, to knockout VP1 for VP1-insertion approaches and VP2 (knockout) for VP-2 fusion. This site-directed mutagenesis has to be performed at several constructs:
- P357 --> pAAV_RC (4fold mutated)
will be performed tomorrow by Bea/Stefan (12.9.10)
- P396 --> pAAV_RC (4fold mutated, ins rep, cap)
will be performed tomorrow by Bea/Stefan (12.9.10)
- P413 --> pAAV_RC (4fold mutated, ins cap)
will be performed tomorrow by Bea/Stefan (12.9.10)
- pAAV_RC (4fold mutated, ins rep, SDM KpnI, inserted “cap”) --> since no Mini-preps have been prepared until now, no plasmid number is available.
dependant on test digestion performed tomorrow (12.9.10), if it works out, it will be performed tomorrow (12.9.10) as well
118. labday 12.09.2010
Mini-Prep and test digestion of pAAV_RC_RepCapIns_SDMKpnI
Investigator: Stefan
Glycerol stocks were prepared:
Mini-Prep was performed according to the standard protocol
Constructs were digested with KpnI for 70 minutes at 37°C, as controls the following constructs were used:
| components | pAAV_RC_RepCapIns_SDMKpnI clone 1-3 | pAAV_RC_1.2SDMSalI (P158) | pAAV_RC_CapIns_prepSDM (P396) | pAAV_RC_CapIns_noRep (P413) |
| DNA | 1 | 1 | 1 | 1 |
| Buffer 1 (10x) | 1 | 1 | 1 | 1 |
| BSA (10x) | 1 | 1 | 1 | 1 |
| KpnI | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 6,5 | 6,5 | 6,5 | 6,5 |
| Total volume | 10 | 10 | 10 | 10 |
Preparation of gel:
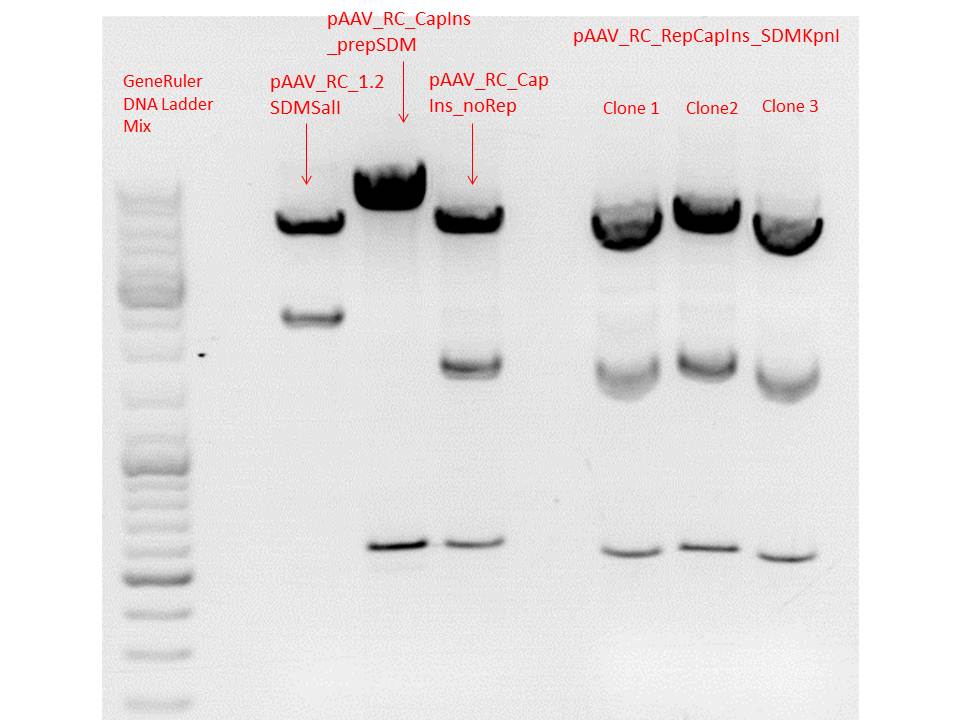
Comment: Test digestions look fine. Work will be continued with clone 2 (P432) which will be sent for sequencing tomorrow as well.
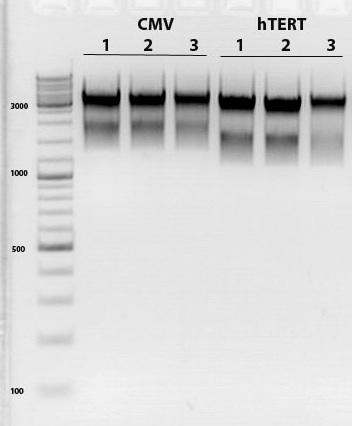
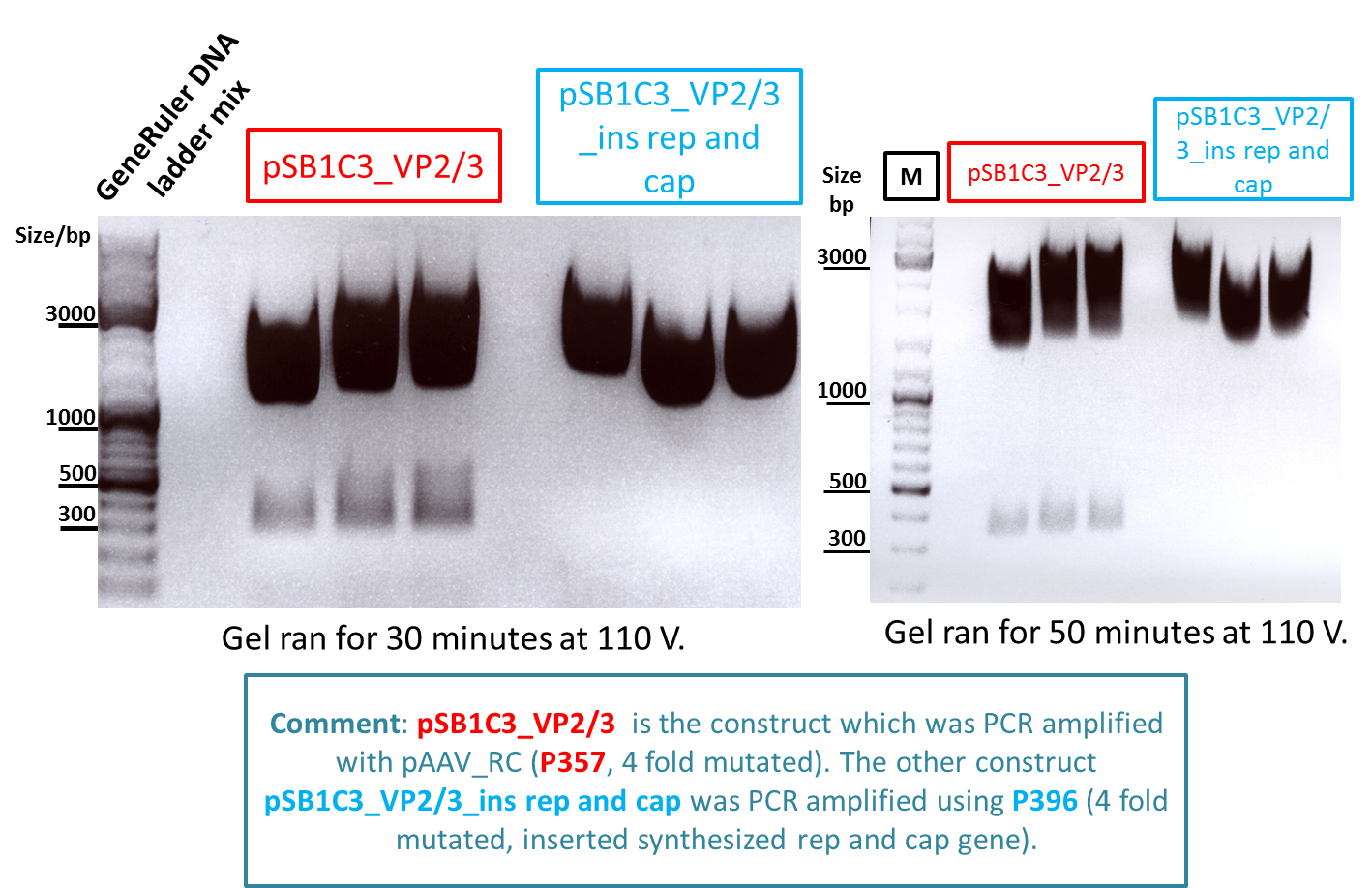
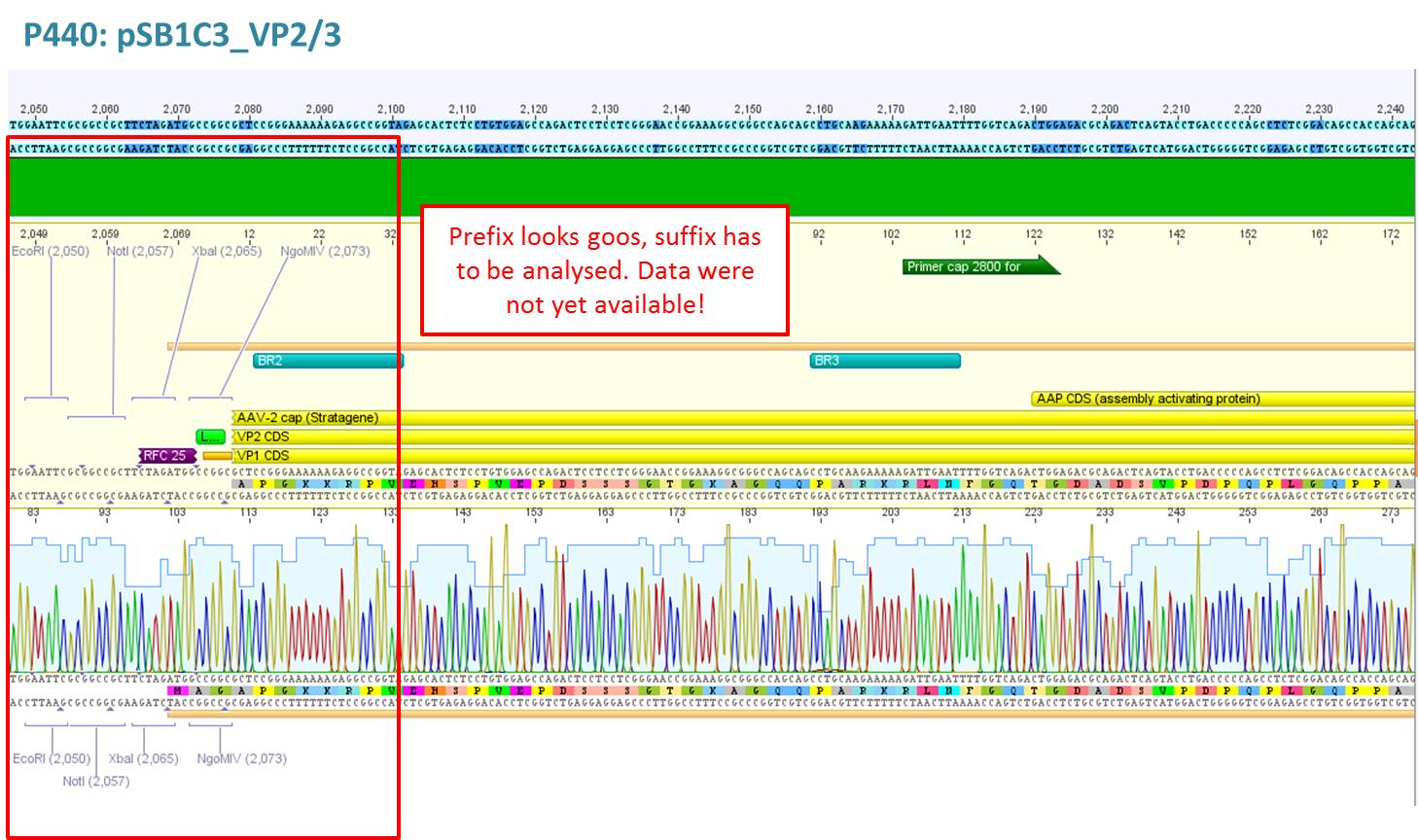
Test digestion of pSB1C3_VP2/3 needed for N-terminal fusion
Investigator: Bea
Comment: Test digestion of the two different approaches Jessica started on Friday has been performed. The general idea of the approach is to obatain VP2/3 cap genes (which means that the 137 amino acids of VP1 are missing) which then can be fused to the targeting molecules for the VP1 insertion and VP3 fusion approaches.
Note: P257_1 - 3 means that the pCR has been carried out with this construct. Same can be pbserved with the plasmid number P296.
Test digestion has been carried out using following enzymes:
- Enzyme 1: PstI-HF
- Enzyme 2: NgomIV
| Mastermix/µL | P357_1/µL | P357_2/µL | P357_2/µL | P396_1/µL | P396_2/µL | P396_2/µL | |
| DNA | - | 2 | 2 | 2 | 2 | 2 | 2 |
| BSA (10x) | - | 2,5 | 2,5 | 2,5 | 2,5 | 2,5 | 2,5 |
| Buffer 4 (10x) | 10,5 | ||||||
| Enzyme NgoMIV | 3,5 | ||||||
| Enzyme PstI | 3,5 | ||||||
| H2O | - | 10,5 | 10,5 | 10,5 | 10,5 | 10,5 | 10,5 |
| Total volume | 15 | 15 | 15 | 15 | 15 | 15 | 15 |
- Test digestion was carried out for 55 minutes at 37°C.
- The samples were loaded on a 1% agarose gel
Loading plan:
M P357_1 P357_2 P357_3 P396_1 P396_2 P396_3
Results:
-
Expected sizes of the two approaches has been:
- "P357" = pSB1C3_RC: 304bp , 1651bp , 2061bp
- "P396"= pSB1C3_RC_ins_rep and cap: 1951bp , 2061bp
Quickchange site directed mutagenesis of pAAV_RC: VP1/VP2 knockout
Investigator: Stefan
Comment: A site-directed mutagenesis has to be performed in order to obtain the plasmids which can be co-transfected with the modified construct containing the targeting molecules. Aim is to knockout VP1 for VP1-insertion approaches and VP2 (knockout) for VP-2 fusion. This site-directed mutagenesis has to be performed at several constructs:
Knock-out of VP1:
Knock-out of VP2:
Used primers:
Knock-out of VP1:
Knock-out of VP2:
Used plasmid and water per approach:
| plasmid no | used plasmid (same for both approaches) (1:100) / µl | VP1-ko: used water / µl | VP2-ko: used water / µl |
| P158 | 0,5 | 18,99 | 19,23 |
| P396 | 2 | 17,49 | 17,73 |
| P413 | 3 | 16,49 | 16,73 |
| P432 | 3 | 16,49 | 16,73 |
Ingredients:
| Ingredients | VP1-ko approach Volume / µl | VP2-ko approach Volume / µl |
| 10x reaction buffer | 2,5 | 2,5 |
| dNTP | 0,5 | 0,5 |
| forward primer: | O170 - 0,38 | O165 - 0,50 |
| reverse primer: | O171 - 0,38 | O166 - 0,52 |
| DNA Template | see above | see above |
| QuikSolution Reagent | 0,75 | 0,75 |
| QuikChangeLightning Enzyme | 0,5 | 0,5 |
| H2O | see above | see above |
| Total volume | 25 | 25 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 1 | 95 | 120 |
| 2 8x(step 2-4) | 95 | 15 |
| 3 | 60 | 10 |
| 4 | 68 | 225 |
| 5 | 68 | 600 |
After PCR, the products were incubated with DpnI at 37 °C for 10 minutes.
Transformation:
Transformation was performed according to standard protocol using XL10-Gold cells. Cells were plated on plate containing Ampicilin.
Comment: There were not enough cells for all approaches. Therefore, only transformation of approach 5 and 8 were performed using 45 µl of cells. The other approaches were conducted with 30 µl.
119. labday 13.09.2010
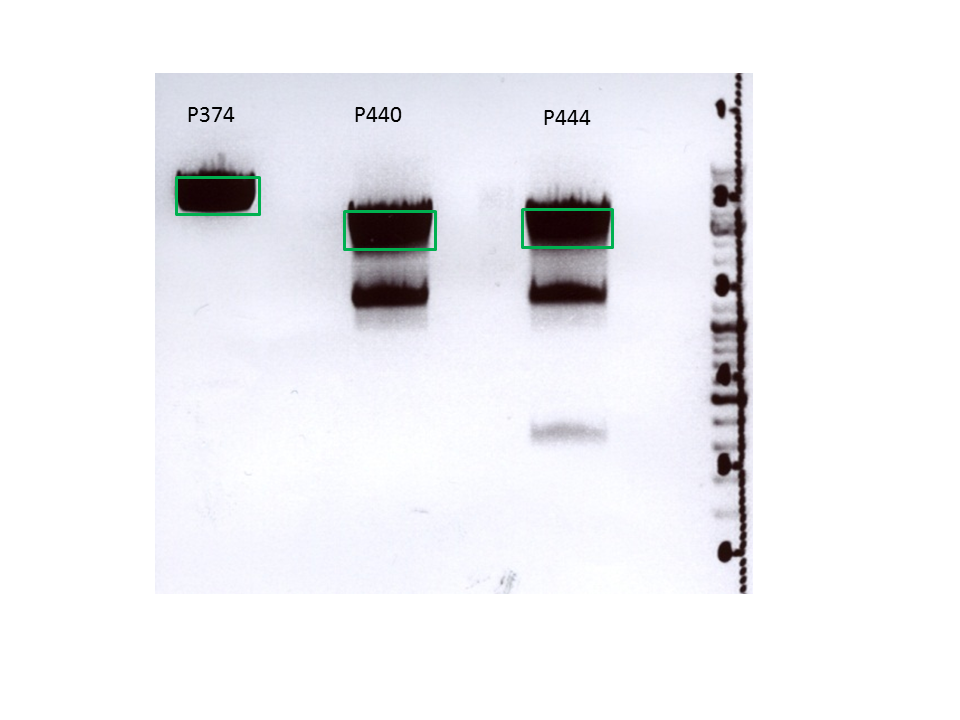
Biobrick assembly: Cloning VP2/3 and VP2/3_ins_Cap to several targeting approaches
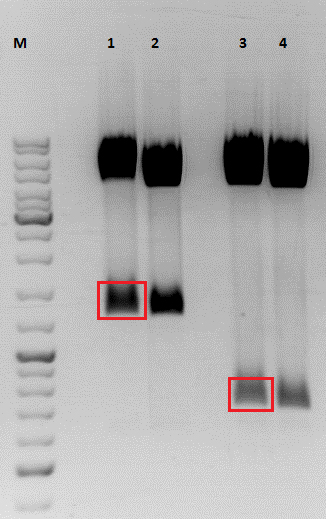
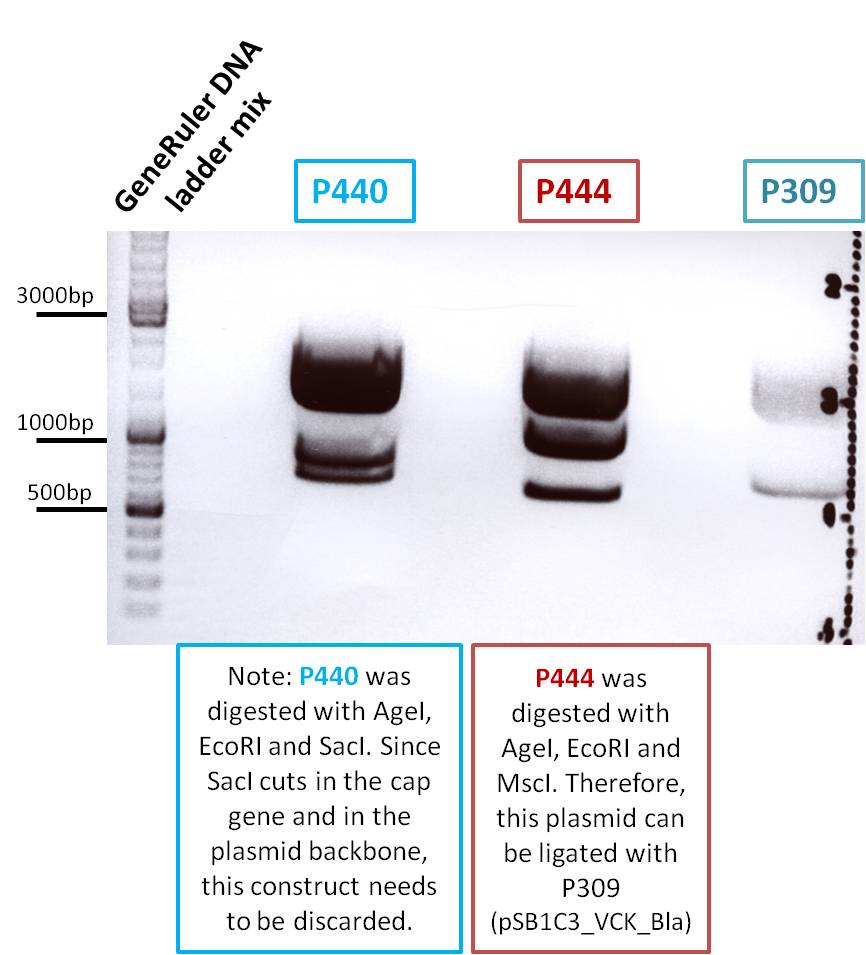
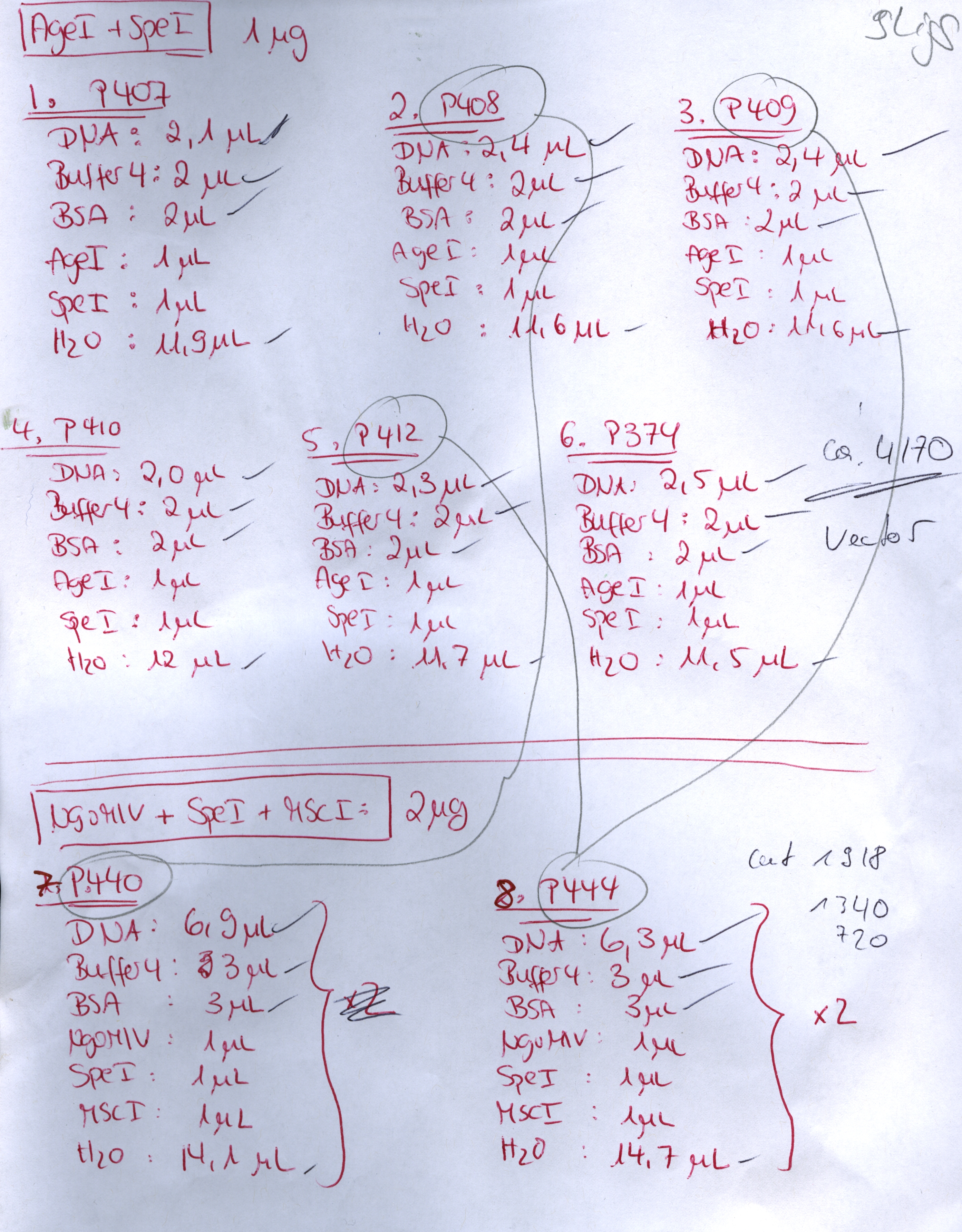
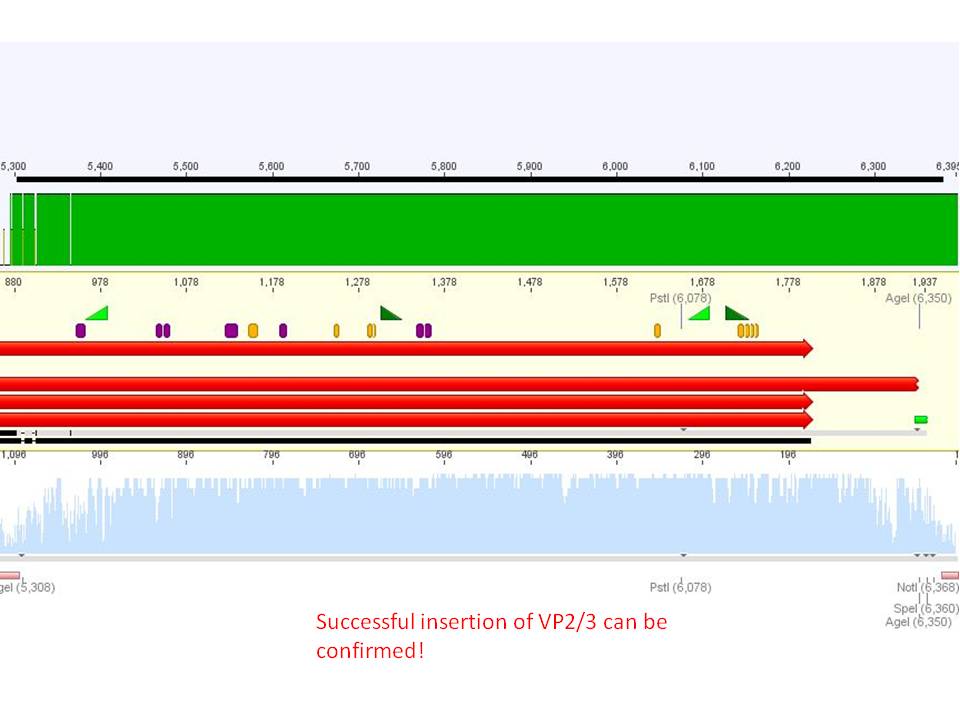
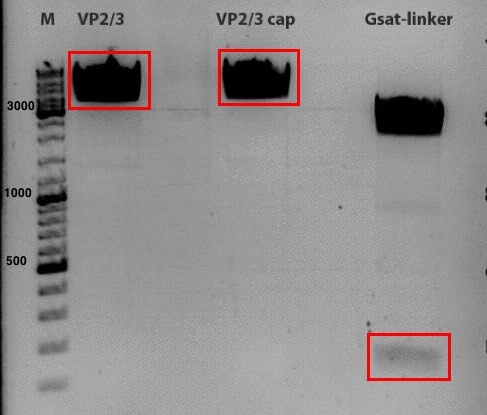
Investigator: Stefan
Comment: Goal of the experiment is to fuse VP2/3 or VP2/3_ins_Cap to pCerulean_VP1up_NLS_Affibody, pCerulean_VP1up_NLS_ Histag and pCerulean_VP1up_NLS_mVenus.
Digestion of the constructs:
Inserts:
- pSB1C3_VP2/3 (P440) c= 290,74 ng/µl
- pSB1C3_VP2/3_ins_cap (P444) c= 314,58 ng/µl
Vectors:
- pCerulean_VP1up_NLS_Affibody (P422) c= 390,68 ng/µl
- pCerulean_VP1up_NLS_Histag (P424) c= 439,57 ng/µl
- pCerulean_VP1up_NLS_mVenus (P426) c= 416,59 ng/µl
Restriction enzymes:
The inserts were digested using SpeI and NgoMIV, the vectors were digested using AgeI and SpeI.
| Components | P440 /µL | P444 /µL | P422 /µL | P424 /µL | P426 /µL |
| DNA | 5 | 4,5 | 6,5 | 5,5 | 6 |
| BSA (10x) | 2 | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 |
| Enzyme 1 | 1 | 1 | 1 | 1 | 1 |
| Enzyme 2 | 1 | 1 | 1 | 1 | 1 |
| H2O | 9 | 9,5 | 7,5 | 8,5 | 8 |
| Total volume | 20 | 20 | 20 | 20 | 20 |
Gel:
0.5 g Agarose, 50 ml TAE (1 %), 3 µL GELRED, at 90 Volt, running time: ~90 minutes
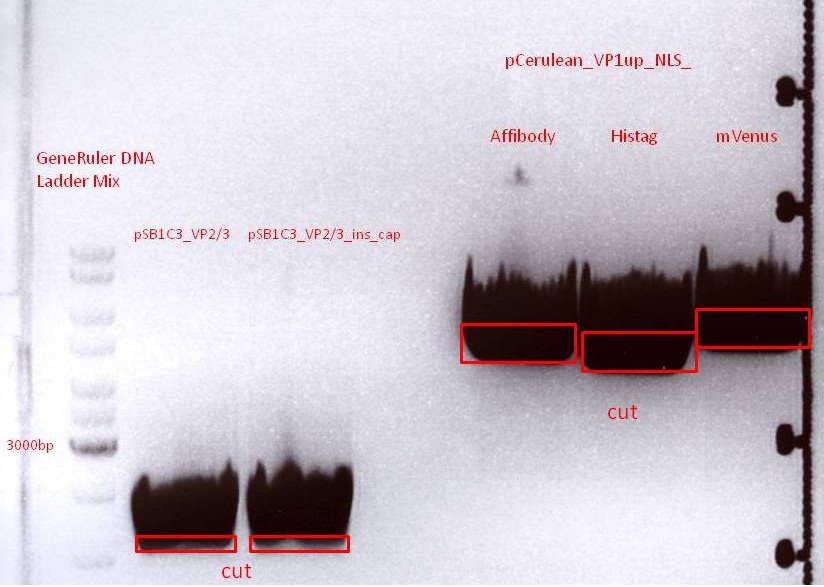
Gel-Extraction:
Marked bands on the gel were cut out. Gel extraction was performed according to protocol.
T4 Ligation:
All ligation approaches were performed using 1 µl of vector and 7 µl of insert. 1 µl of T4 ligation buffer and 1 µl of T4 ligase were used.
Transformation:
Transformation was performed according to standard protocol using BL21 cells and plates containing Kanamycin.
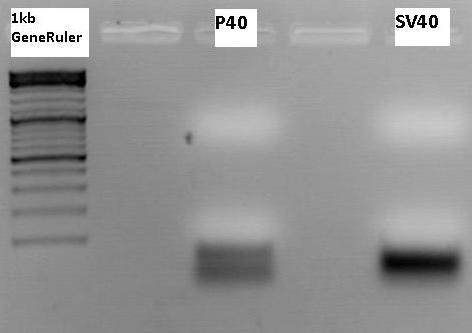
Biobrick production: P40 & SV40
Investigator: Patrick
P158 (398 ng/µl) was used as P40 template and P230 (367 ng/µl) as SV40 template. Both of them were diluted 1:400.
SV40:
| Ingredients | volume |
| 5X Phusion HF buffer | 10 µl |
| 10 mM dNTP mix | 1µl |
| forward primer: O158 | 2,5 µl |
| reverse primer: O159 | 2,5 µl |
| DNA Template | 1 µl |
| DMSO | 0 µl |
| Phusion Polymerase | 0,5 µl |
| H2O | 32,5 µl |
| Total volume | 50 µl |
| Cycles | Temperature | Time |
| 98°C | 1 | |
| 8x | 98°C | 15" |
| 59°C | 25" | |
| 72°C | 7" | |
| 17x | 98°C | 15" |
| 67°C | 25" | |
| 72°C | 7" | |
| 1x | 72°C | 5' |
| Hold 4°C |
P40:
| Ingredients | Volume / µl |
| 5X Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| forward primer: O152 | 2,5 |
| reverse primer: O153 | 2,5 |
| DNA Template | 1 |
| DMSO | - |
| Phusion Polymerase | 0,5 |
| H2O | 32,5 |
| Total volume | 50 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 98 | 60 | |
| 8x | 98 | 15 |
| 61 | 25 | |
| 72 | 10 | |
| 17x | 98 | 15 |
| 69 | 25 | |
| 72 | 10 | |
| 1x | 72 | 300 |
| Hold | 4 |
Expected size of the PCR products: P158/p40: 180 bp, P230/SV40: 230 bp.
The visible bands do not have the expected size. These are probably amplified primers. The experiment has to be repeated.
Tobi proposed the following changes:
Use 1,5 ng template for p40 and SV40.
p40 PCR program:
| Cycles | Temperature / °C | Time / s |
| 98 | 30 | |
| 10x | 98 | 10 |
| 58 | 25 | |
| 72 | 10 | |
| 15x | 98 | 10 |
| 66 | 25 | |
| 72 | 10 | |
| 1x | 72 | 300 |
| Hold | 4 |
SV40 PCR program:
| Cycles | Temperature | Time |
| 98°C | 30" | |
| 10x | 98°C | 10" |
| 56°C | 25" | |
| 72°C | 10" | |
| 15x | 98°C | 10" |
| 64°C | 25" | |
| 72°C | 10" | |
| 1x | 72°C | 5' |
| Hold 4°C |
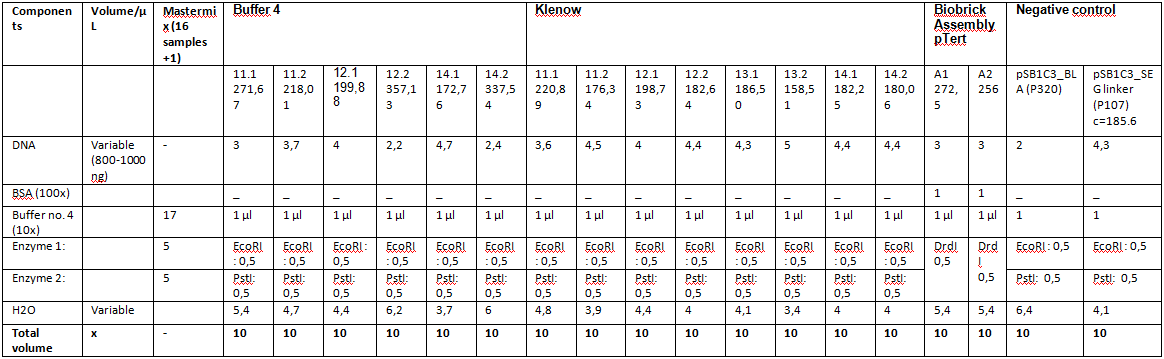
Test digestion of ViralBrick ligation and pTert BioBrick assembly
Investigator: Achim, Anna
- Our Samples were digested with EcoRI and PstI again.
- We used negative controls pSB1C3_Bla, containing a ~800bp insert and pSB1C3_SEG_linker, containing a ~160bp insert.
- Additionally, a new test digestion of the pTert Biobrick assembly was prepared,this time with DrdI, an enzyme that should cut once in the insert and once in the vector. Expected sizes: ~2000 & 2500bp
- Test Digestion ViralBricks:
- Test Digestion Assembly
PCR of RepCap/Cap
Investigator: Jessica
DNA sample was diluted 1:100
- P158 = pAAV_RC_1.2 SDM SalI
- P396 = pAAV_RC_CapIns_prepSDM clone 3
- P413 = pAAV_RC_CapIns_noRep clone 1
- P432 = pAAV_RC_RepCapIns_SDMKpnI clone 2
PCR progam:
for Rep Cap
| Ingredients | Volume P158 / µl | Volume P396/ µl | Volume P413/ µl | Volume P432/ µl |
| 5X Phusion HF buffer | 10 | 10 | 10 | 10 |
| 10 mM dNTP mix | 1 | 1 | 1 | 1 |
| forward primer: O93 | 2,5 | 2,5 | 2,5 | 2,5 |
| reverse primer: O120 | 2,5 | 2,5 | 2,5 | 2,5 |
| DNA Template | 0,25 | 0,20 | 0,32 | 0,30 |
| DMSO | - | - | - | - |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 33,25 | 33,30 | 33,18 | 33,20 |
| Total volume | 50 | 50 | 50 | 50 |
no DNA on the gel...
for Cap
| Ingredients | Volume P158 / µl | Volume P413/ µl |
| 5X Phusion HF buffer | 10 | 10 |
| 10 mM dNTP mix | 1 | 1 |
| forward primer: O89 | 2,5 | 2,5 |
| reverse primer: O120 | 2,5 | 2,5 |
| DNA Template | 0,25 | 0,32 |
| DMSO | - | - |
| Phusion Polymerase | 0,5 | 0,5 |
| H2O | 33,25 | 33,18 |
| Total volume | 50 | 50 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 98 | 60 | |
| 8x | 98 | 15 |
| 60 | 25 | |
| 72 | 70 | |
| 17x | 98 | 15 |
| 69 | 25 | |
| 72 | 70 | |
| 1x | 72 | 300 |
| Hold | 4 |
Digestion of plasmid backbone:
Plasmid used: pSB1C3_VCK_Bla (P320)
c (pSB1C3_VCK_Bla) = 408,0 ng/ µl
| Components | vector Volume/µL |
| DNA | 2,45 |
| BSA (10x) | 2 |
| Buffer no. 4 (10x) | 2 |
| NgoMIV | 1 |
| SpeI | 1 |
| H2O | 11,55 |
| Total volume | 20 |
incubation @ 37 C for approx. 2h
1% agarose gel
Digestion of PCR product:
| Components | PCR product Volume/µL |
| DNA | 30 |
| BSA (10x) | 4 |
| Buffer no. 4 (10x) | 4 |
| NgoMIV | 1 |
| SpeI | 1 |
| H2O | - |
| Total volume | 40 |
incubation @ 37 C for approx. h
Ligation:
1 µl T4 DNA ligase, 1 µl 10x Buffer, µl vector (P320), µl Insert (P158)/ µl Insert (P413).
Incubation time:
Miniprep and test digestion of CD biobricks
Investigator: Kira
| Components | sample Volume/µL |
| DNA | 4 |
| BSA (10x) | 2 |
| Buffer no. 4 (10x) | 2 |
| XbaI | 1 |
| AgeI-HF | 0,5 |
| H2O | 10,5 |
| Total volume | 20 |
incubation @ 37 C for approx. 2h
1% agarose gel
BioBrick TM assembly of pSB1C3_leftITR_promoter_betaglobin to several GOIs
Investigator: Bea
Comment: Since we only assembled the vecotrplasmid with the CMV promoter and mVenus_YFP as gene of interest other approaches have to be conducted, especially the vectorplasmid which contains the tumorspecific promoter phTERT with the gene of interest mGMK_TK30. These approaches were started by Kira some days ago, and will now be continued. This time we are fusing the extisting and confirmed constructs pSB1C3_leftITR_CMV_betaglobin and pSB1C3_leftITR_phTERT_betaglobin to the gene of interests mVenus_YFP and mGMK.
Digestion of the constructs:
- P434 = pSB1C3_leftITR_CMV_betaglobin c=331ng/µL
- P434 = pSB1C3_leftITR_phTERT_betaglobin c=331ng/µL
| Components | vP434 /µL | vP437 /µL |
| DNA | 4,5 | 4,5 |
| BSA (10x) | 2 | 2 |
| Buffer no. 4 (10x) | 2 | 2 |
| SpeI | 1 | 1 |
| PstI-HF | 1 | 1 |
| H2O | 9,5 | 9,5 |
| Total volume | 20 | 20 |
Digestion of the three targeting approaches:
- P81 = pAAV_iGEM_mGMK_TK30 --> approach will not be proceeded becuase the iGEM restriction sites cut in the vctor as well and therefore the desired fragments cannot be isolated with used iGEM restriction sites.!!!
- P195 = pSB1C3_mGMK c=258ng/µL
- P229 = pAAV_iGEM_mVenus c=486 ng/µL
| Components | vP81 /µL | vP195 /µL | vP229 /µL |
| DNA | 7,8 | 12,3 | 4,1 |
| BSA (10x) | 2,5 | 2,5 | 2,5 |
| Buffer no. 4 (10x) | 2,5 | 2,5 | 2,5 |
| XbaI | 1 | 1 | 1 |
| PstI-HF | 1 | 1 | 1 |
| H2O | 10,2 | 12,3 | 13,9 |
| Total volume | 25 | 25 | 25 |
Loading plan:
M P434 P437 P81 P195 P229
Results of preparative agarose gel:
Expected sizes:
-
P434 = 3300bp
P437 = 3183 bp
P195 = 627bp, 2054bp
P229 = 756bp, 667bp, 2600bp, 1300bp
After the gel extraction (the fragments cut out of the gel were labeled) has been performed the ligation was carried out.
Ligation:
- v=P434 =5,61µL
- v=P195 =2,39µL
- v=P437 =4,77µL
- v=P195 =3,32µL
- v=P437 =4,17µL
- v=P229 =3,83µL
After mixing insert and vector into a tube, add 1µL T4-ligase 10x buffer and 1µL ligase to the DNA mix and incubate for 40 minutes at room temperature.
Transform XL1-B cells, and plate them on agar containing chloramphenicol.
Next steps: Picking clones of the constructs, prepare Mini-prep and finally fuse them to the existing construct pSB1C3_hGH_rightITR!
Sent the construct pSB1C3_VP2/3 and pSB1C3_VP2/3 for sequencing
Investigator: Bea
Comment: The construct which has been produced (experiment started on friday) will be sequenced today. The construct were named: P357 1- 3 and P396 1-3. After preparing for sequencing the plasmids were renamed and received a plasmid number aswell.
- P357_1 = P440
- P357_2 = P441
- P357_3 = P442
- P396_1 = P443
- P396_3 = P443
- P396_3 = P445
The two constructs which have been sent for sequencing are:
- P440 = BK_1, used primers: VR-2 and VF-2
- P444 = BK_2, used primers: VR-2 and VF-2
Fusion of VP2/3 to C-terminus of Zegfr:1907_"Linker", CFP_MiddleLinker and 6xHis_MiddleLinker construct
Investigator: Hanna
UPDATE: The Zegfr:1907 Affibody was successfully fused to the different linker (Short Linker, Long Linker, Middle Linker and SEG Linker) in order to find out with which linker targeting functions best. In addition to that, CFP (Imaging) and the His-Tag (=6xHis, Purification) was fused to the Middle Linker. These constructs were cloned into the pCerulean plasmid (in order to be driven by the CMV promoter + SV40 terminator).
The last steps in the VP2-N-terminal fusion approach include the knockout of the VP2 start codon in pAAV_RC (different constructs: 4-fold mutated, 4-fold mutated with Cap-insert, 4-fold mutated with Rep/Cap-insert/KpnI-backmutation) and fusion of the VP2/3 ORF to the C-terminus of the linker.
SDM of the VP2 start codon (of the unmodified and Cap-insert sequence) was already performed by Stefan. After the Cap-insert variant has been tested in cell culture, the HSPG-KnockOut motif will be cloned into Cap and also used for BioBrick production via PCR. This fragment will also be used for fusion to the C-terminus of the Linker-Library+Affibody and the purification (6xHis) and Imaging (CFP, better: GFP) approaches. Therefore pAAV_RC_Rep/Capins_KpnI-backmutation_HSPG-KO (if Rep-KpnI-backmutation works!) will be also used for SDM of VP2 start codon.
Today VP2/3 (unmodified) and VP2/3-Cap-ins will be fused to the C-terminus of the linker. These approaches will be co-transfected with the referring pAAV_RC_VP2-KO constructs and therefore tested in cell culture on wednesday.
Practical Cloning:
- Vector: pCerulean_CFP_MiddleLinker (P407), pCerulean_Zegfr:1907_MiddleLinker (P408), pCerulean_Zegfr:1907_SEG (P409), pCerulean_Zegfr:1907_ShortLinker (P410), pCerulean_Zegfr:1907_LongLinker (P412), pCerulean_6xHis_MiddleLinker (P374)
- Insert: pSB1C3_VP2/3 (P440), pSB1C3_VP2/3_ins_cap (P444)
- new vector name: pCerulean_CFP_MiddleLinker_VP2/3, pCerulean_Zegfr:1907_MiddleLinker_VP2/3, pCerulean_Zegfr:1907_SEG_VP2/3, pCerulean_Zegfr:1907_ShortLinker_VP2/3, pCerulean_Zegfr:1907_LongLinker_VP2/3, pCerulean_6xHis_MiddleLinker_VP2/3 AND pCerulean_CFP_MiddleLinker_VP2/3-insCap, pCerulean_Zegfr:1907_MiddleLinker_VP2/3-insCap, pCerulean_Zegfr:1907_SEG_VP2/3-insCap, pCerulean_Zegfr:1907_ShortLinker_VP2/3-insCap, pCerulean_Zegfr:1907_LongLinker_VP2/3-insCap, pCerulean_6xHis_MiddleLinker_VP2/3-insCap
- buffer used:4
Comment Because the VP2/3 frament has approximately the same size as the backbone pSB1C3 it may be difficult to separate them on the agarose gel. Therefore a single cutter enzyme (SacI) was chosen to digests the backbone. This enzyme also works in buffer 4 + BSA and needs to be incubated at 37°C.
- Vector: AgeI-HF, SpeI
- Insert: NgoMIV, SpeI, SacI
Digestion
| components | volume of vector /µl | volume of insert /µl |
| DNA | 3.75 | 8.3 |
| BSA (10x) | 2 | 3 |
| Buffer 4(10x) | 2 | 3 |
| AgeI-HF | 1 | - |
| NgoMIV | - | 1 |
| SpeI | 1 | 1 |
| SacI | - | 1 |
| H2O | 10.25 | 12.7 |
| Total volume | 20 | 30 |
- Incubation: 2 h
Comment_ Because we are all hardworking cloners :) there was not enough AgeI left for all 6 reactions (just 3: P374, P407, P408). Unfortunately the lab stock AgeI was also empty. Therefor we had to "lend" it from the Baumeister group - which is already from "Fermentas" and therefore also needs Fermentas-buffer. Nevertheless we checked the pH from buffer 4 with the one needed for the Fermentas-AgeI and decided to try using this AgeI in buffer 4.
Agarose-Gel:
0.8 g and 0.4 g Agarose, 50 mL TAE (0.8 % and 1.5 %), 3 µL GELRED, at 90 Volt, running time:60 minutes
Comment: Unfortunately the gels were interchanged so that the vector was loaded onto the 1.5% gel.
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 | Expected size 2 | Expected size 3 |
|---|---|---|---|---|---|
| P407 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P408 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P409 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P410 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P412 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P374 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P440 | 30 µl | 6 µl | 1900 bp | 910 bp | 1150 bp |
| P444 | 30 µl | 6 µl | 1900 bp | 910 bp | 1150 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample Jessy /µl | Sample P407 /µl | Sample P408 /µl | Sample P409 /µl | Sample P410 /µl | Sample P412 /µl | |
|---|---|---|---|---|---|---|---|
| Lane | 5.5 | 24 | 24 | 24 | 24 | 24 |
| Sample P374 /µL | Sample P440 /µl | Sample P444 /µl | Marker /µl | |
|---|---|---|---|---|
| Lane | 24 | 36 | 36 | 5.5 |
Gel extraction
Gel measurement:
| Sample | Weight |
| P407 | 150 mg |
| P408 | 180 mg |
| P409 | 230 mg |
| P410 | 220 mg |
| P412 | 170 mg |
| P374 | 180 mg |
| P440 | 210 mg |
| P444 | 180 mg |
Ligation
| Construct | Vector (µl) | Insert (µl) |
| 407 + 440 | 4.6 | 3.4 |
| 407 + 440 | 4.98 | 3.02 |
| 408 + 440 | 4.14 | 3.86 |
| 408 + 444 | 4.53 | 3.47 |
| 409 + 440 | 4.18 | 3.82 |
| 409 + 444 | 4.57 | 3.43 |
| 410 + 440 | 3.6 | 4.4 |
| 410 + 444 | 3.99 | 4.01 |
| 412 + 440 | 3.6 | 4.4 |
| 412 + 444 | 3.99 | 4.01 |
| 374 + 440 | 3.23 | 4.77 |
| 374 + 444 | 3.61 | 4.39 |
120. labday 14.09.2010
Test Digestion of remaining ViralBrick clones
Investigator: Achim
ViralBrick sequencing results
Investigator: Achim
- The two plasmids contain Z34C, but are incomplete in the 453 upstream region. There could be several reasons for this: Incompletely synthesized oligos, incomplete fill-in reactions...
- Tomorrow we're going to try another repetition of the hybridisation and ligation, this time with dephosohorized vector - that should prevent the growth of clones containing religated vector. We will also use new Klenow fragment and try two parallel ligations: Quickligation & T4 ligation over night.
Retrafo with sr39_rightITR (Mr.Gene)
Investigator: Bea
Comments: We received the sr39 (mutant verstion of the wt thymidinkinase) and wt rightITR ordered from Mr. Gene today. In order to use and clone the insert into the desired sequences a retrafo has to be performed.
Re-transformation:
- Construct used:
- We reveived 5 µg (lyophilzied). These were resuspended in 100 µL H2O and vortexted.
- c = 50 ng/µL
- c = Plasmid need to be diluted in order to obatain the final 500pg
- Dilution: 1:100
- 100 µL aliquot of XL1-B were thawed on ice. The proper amount of 1µL was added to the cells.
- Transformed cells were plated on LB plates containing kanamycin
Picking clones of pSB1C3_leftITR_promoter_betaglobin_GOI
Investigator: Bea
Comments: Clones were picked from the approach condutced yesterday.
- pSB1C3_leftITR_CMV_betaglobin_mGMK
- pSB1C3_leftITR_phTERT_betaglobin_mGMK
- pSB1C3_leftITR_phTERT_betaglobin_mVenus
Additionally, pHelper (Glycerol stock) was inoculated in 30 mL containing ampicillin.
3rdrepetition of SV40 terminator (hopefully the last one ;-))
Investigator: Bea
Comments: The last repetition of the PCR for obtaining the SV40 fragment did not work out because wrong primers were used therefore the PCR will be carried out once again.
Preparations:
A 1:1000 dilution of the plasmid P230 (pEGFP_C1) which contains the desired SV40 polyadenlyation site will be prepared. This fragment will be extracted and cloned into pSB1C3 iGEM standard plasmid.
PCR protocol:
| Ingredients | volume |
| 5X Phusion HF buffer | 10 µl |
| 10 mM dNTP mix | 1µl |
| forward primer: O158 | 2,5 µl |
| reverse primer: O159 | 2,5 µl |
| DNA Template | 3,7 µl |
| DMSO | 0 µl |
| Phusion Polymerase | 0,5 µl |
| H2O | 29,8 µl |
| Total volume | 50 µl |
The following PCR programm was used. Idea is that in the first 8 cycles the primer can only bind to the template added in to the reaction mix. The following 17 cycles afterwards allow the annealing of the complete primer containing prefix and suffix with its restriciton sites.
| Cycles | Temperature | Time |
| 98°C | 1 | |
| 8x | 98°C | 15" |
| 59°C | 25" | |
| 72°C | 7" | |
| 17x | 98°C | 15" |
| 67°C | 25" | |
| 72°C | 7" | |
| 1x | 72°C | 5' |
| Hold 4°C |
The PCR product (after PCR purification) and the plasmid vector pSB1C3_CFP (P51.2) will be digested with the same restriction enzymes and afterwards loaded on a 1% agarose gel:
- XbaI
- PstI
| Components | vPCR product SV40 /µL | vP51.2 /µL |
| DNA | 20 | 9 |
| BSA (100x) | 0,3 | 0,2 |
| Buffer no. 4 (10x) | 3 | 2 |
| SpeI | 1 | 1 |
| PstI-HF | 1 | 1 |
| H2O | 4,7 | 6,8 |
| Total volume | 30 | 20 |
After digestion of the two constructs they were loaded on a 1% agarose gel.
M PCR product SV40 pSB1C3_CFP
Expected sizes of the two digested constructs are:
- PCR product SV40 terminator: 255bp
- pSB1C3: 2053bp
- pSB1C3: 756bp
Results:
After gel extraction has been performed, the ligation was carried out.
Ligation:
- v=pSB1C3 =7,7µL
- v=SV40 =0,3µL
The ligation mix was transformed into XL1-Blue cells and plated on agar plates containing chloramphenicol.
Next steps:
Picking clones and perform Mini-Prep. This construct can then be submitted to the parts registry after sequencing.
Sequencing analysis from pSB1C3_VP2/3 and pSB1C3_VP2/3_ins_cap
Investigator: Bea
Comment: The constructs which were sent for sequencing are the plasmids which were PCR amplified from the pAAV_RC and cloned into the pSb1C3. Purpose of the PCR amplification was to clone the fragement to the N-terminal targeting approaches.
- P440 = pSB1C3_VP2/3 = BK_1
- Used primers:
- VR-2
- VF-2
Results:
- P444 = pSB1C3_VP2/3_ins_cap = BK_2
- Used primers:
- VR-2
- VF-2
Results:
Midiprep of pAAV_RC_CapIns_NoRep; pAAV_RC_RepCapIns_SDMKpnI
Investigator: Chris W.
The Midiprep was performed according to the standard protocol yieldung the following concentrations:
pAAV_RC_CapIns_NoRep: 387,05 ng/µl, labeled as P467
pAAV_RC_RepCapIns_SDMKpnI: 1157.58 ng/µl labeled as P468
Sequencing analysis from pAAV_RC_RepCapIns_SDMKpnI
Investigator: Stefan
Comment: Last week's SDM to remutate the KpnI restriction site in Rep needed to be sequenced.
- P432 = pAAV_RC_RepCapIns_SDMKpnI = AM_3
- Used primers: 1250 for
Results:
Repetition of the fusion of VP2/3 to C-terminus of Zegfr:1907_"Linker", CFP_MiddleLinker and 6xHis_MiddleLinker construct
Investigator: Anissa
COMMENT: The same approach which has been done yesterday, was repeated today, because gel-pictures showed no satisfying results.
Practical Cloning:
- Vector: pCerulean_CFP_MiddleLinker (P407), pCerulean_Zegfr:1907_MiddleLinker (P408), pCerulean_Zegfr:1907_SEG (P409), pCerulean_Zegfr:1907_ShortLinker (P410), pCerulean_Zegfr:1907_LongLinker (P412), pCerulean_6xHis_MiddleLinker (P374)
- Insert: pSB1C3_VP2/3 (P440), pSB1C3_VP2/3_ins_cap (P444)
- new vector name: pCerulean_CFP_MiddleLinker_VP2/3, pCerulean_Zegfr:1907_MiddleLinker_VP2/3, pCerulean_Zegfr:1907_SEG_VP2/3, pCerulean_Zegfr:1907_ShortLinker_VP2/3, pCerulean_Zegfr:1907_LongLinker_VP2/3, pCerulean_6xHis_MiddleLinker_VP2/3 AND pCerulean_CFP_MiddleLinker_VP2/3-insCap, pCerulean_Zegfr:1907_MiddleLinker_VP2/3-insCap, pCerulean_Zegfr:1907_SEG_VP2/3-insCap, pCerulean_Zegfr:1907_ShortLinker_VP2/3-insCap, pCerulean_Zegfr:1907_LongLinker_VP2/3-insCap, pCerulean_6xHis_MiddleLinker_VP2/3-insCap
- buffer used:4
Comment Because the VP2/3 frament has approximately the same size as the backbone pSB1C3 it may be difficult to separate them on the agarose gel. Therefore a 0,8% gel was prepared and the samples run for 1,5 h. So the two bands (1900bp and 2200bp) will hopefully be seperated.
- Vector: AgeI-HF, SpeI
- Insert: NgoMIV, SpeI
Digestion
| components | pSb1C3_Vp2/3 (2,5µg) P440 /µl /µl | pSb1C3_Vp2/3_capIns (2,5µg) P444 /µl |
| DNA | 8,6 | 8 |
| BSA (10x) | 1,5 | 1,5 |
| Buffer 4(10x) | 1,5 | 1,5 |
| speI | 1 | 1 |
| NgoMIV | 1 | 1 |
| SpeI | 1 | 1 |
| H2O | 1,4 | 2 |
| Total volume | 15 | 15 |
| components | pCerulean_affib._shortlinker P410 (1µg)/µl | pCerulean_affib._middlelinker P408 (1µg)/µl |
| DNA | 2 | 2,5 |
| BSA (10x) | 1,5 | 1,5 |
| Buffer 4(10x) | 1,5 | 1,5 |
| speI | 1 | 1 |
| AgeI | 0,75 | 0,75 |
| SpeI | 1 | 1 |
| H2O | 8,25 | 7,75 |
| Total volume | 15 | 15 |
| components | pCerulean_affib._longlinker P412 (1µg)/µl | pCerulean_affib._SEG P409 (1µg)/µl |
| DNA | 2,4 | 2,4 |
| BSA (10x) | 1,5 | 1,5 |
| Buffer 4(10x) | 1,5 | 1,5 |
| speI | 1 | 1 |
| AgeI | 0,75 | 0,75 |
| SpeI | 1 | 1 |
| H2O | 7,85 | 7,85 |
| Total volume | 15 | 15 |
| components | pCerulean_CFP_middlelinker P407 (1µg)/µl | pCerulean_His-tag._middlelinker P374 (1µg)/µl |
| DNA | 2,1 | 2,5 |
| BSA (10x) | 1,5 | 1,5 |
| Buffer 4(10x) | 1,5 | 1,5 |
| speI | 1 | 1 |
| AgeI | 0,75 | 0,75 |
| SpeI | 1 | 1 |
| H2O | 8,15 | 7,84 |
| Total volume | 15 | 15 |
- Incubation: 1,5 h
Comment: 0,75 µl AgeI were used because AgeI is not deluted.
Agarose-Gel:
0.4 g and 0,5g Agarose, 50 mL TAE (0.8 %, 1%), 3 µL GELRED, at 110 Volt, running time:1,5 h
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 | Expected size 2 | Expected size 3 |
|---|---|---|---|---|---|
| P407 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P408 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P409 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P410 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P412 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P374 | 20 µl | 4 µl | ~ 4170 bp | - | - |
| P440 | 30 µl | 6 µl | 1900 bp | 910 bp | 1150 bp |
| P444 | 30 µl | 6 µl | 1900 bp | 910 bp | 1150 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample p410 /µl | Sample P408 /µl | Sample P412 /µl | Sample P409 /µl | Sample P407 /µl | Sample P374 /µl | |
|---|---|---|---|---|---|---|---|
| Lane | 3 | 19 | 19 | 19 | 19 | 19 | 19 |
| Marker /µl | Sample P444 /µl | Sample P440 /µl
| |
|---|---|---|---|
| Lane | 3 | 19 | 19 |
Gel extraction
Gel measurement:
| Sample | Weight |
| P407 | 180 mg |
| P408 | 160 mg |
| P409 | 90 mg |
| P410 | 190 mg |
| P412 | 230 mg |
| P374 | 190 mg |
| P440 | 120 mg |
| P444 | 130 mg |
Ligation
| Construct | Vector (µl) | Insert (µl) |
| 407 + 440 | 1 | 3 |
| 407 + 440 | 1 | 3 |
| 408 + 440 | 1 | 3 |
| 408 + 444 | 1 | 3 |
| 409 + 440 | 1 | 3 |
| 409 + 444 | 1 | 3 |
| 410 + 440 | 1 | 3 |
| 410 + 444 | 1 | 3 |
| 412 + 440 | 1 | 3 |
| 412 + 444 | 1 | 3 |
| 374 + 440 | 1 | 3 |
| 374 + 444 | 1 | 3 |
Comment: Because the amount of the insert-DNA wouldn't sufficiend for all ligations, the amount of the totl DNA was reduced to 4 µl.
Transformation
Transformation was performed in Xl1 blue according the standard-protocol.
Mini-Prep and test digestion of multiple plasmids
Investigator: Stefan
Glycerol stocks were prepared:
- B382 = pAAV_RC_1.2SDMSalI_VP1-ko clone 1
- B383 = pAAV_RC_1.2SDMSalI_VP1-ko clone 2
- B384 = pAAV_RC_CapIns_prepSDM_VP1-ko clone 1
- B385 = pAAV_RC_CapIns_prepSDM_VP1-ko clone 2
- B386 = pAAV_RC_CapIns_noRep_VP1-ko clone 1
- B387 = pAAV_RC_CapIns_noRep_VP1-ko clone 2
- B388 = pAAV_RC_RepCapIns_SDMKpnI_VP1-ko clone 1
- B389 = pAAV_RC_RepCapIns_SDMKpnI_VP1-ko clone 2
- B390 = pAAV_RC_1.2SDMSalI_VP2-ko clone 1
- B391 = pAAV_RC_1.2SDMSalI_VP2-ko clone 2
- B392 = pAAV_RC_CapIns_prepSDM_VP2-ko clone 1
- B393 = pAAV_RC_CapIns_prepSDM_VP2-ko clone 2
- B394 = pAAV_RC_CapIns_noRep_VP2-ko clone 1
- B395 = pAAV_RC_CapIns_noRep_VP2-ko clone 2
- B396 = pAAV_RC_RepCapIns_SDMKpnI_VP2-ko clone 1
- B397 = pAAV_RC_RepCapIns_SDMKpnI_VP2-ko clone 2
- B398 = pCerulean_VP1up_NLS_Zegfr:1907_VP2/3 clone 1
- B399 = pCerulean_VP1up_NLS_Zegfr:1907_VP2/3 clone 2
- B400 = pCerulean_VP1up_NLS_Zegfr:1907_VP2/3-Capins clone 1
- B401 = pCerulean_VP1up_NLS_Zegfr:1907_VP2/3-Capins clone 2
- B402 = pCerulean_VP1up_NLS_6xHis_VP2/3 clone 1
- B403 = pCerulean_VP1up_NLS_6xHis_VP2/3 clone 2
- B404 = pCerulean_VP1up_NLS_6xHis_VP2/3-Capins clone 1
- B405 = pCerulean_VP1up_NLS_6xHis_VP2/3-Capins clone 2
- B406 = pCerulean_VP1up_NLS_mVenus_VP2/3 clone 1
- B407 = pCerulean_VP1up_NLS_mVenus_VP2/3 clone 2
- B408 = pCerulean_VP1up_NLS_mVenus_VP2/3-Capins clone 1
- B409 = pCerulean_VP1up_NLS_mVenus_VP2/3-Capins clone 2
Mini-Prep was performed according to the standard protocol
- P449 = pAAV_RC_1.2SDMSalI_VP1-ko clone 1 c= 372,90 ng/µl
- P450 = pAAV_RC_1.2SDMSalI_VP1-ko clone 2 c= 380,47 ng/µl
- P451 = pAAV_RC_CapIns_prepSDM_VP1-ko clone 1 c= 367,08 ng/µl
- P452 = pAAV_RC_CapIns_prepSDM_VP1-ko clone 2 c= 387,34 ng/µl
- P453 = pAAV_RC_CapIns_noRep_VP1-ko clone 1 c= 360,67 ng/µl
- P454 = pAAV_RC_CapIns_noRep_VP1-ko clone 2 c= 353,62 ng/µl
- P455 = pAAV_RC_RepCapIns_SDMKpnI_VP1-ko clone 1 c= 353,85 ng/µl
- P456 = pAAV_RC_RepCapIns_SDMKpnI_VP1-ko clone 2 c= 349,39 ng/µl
- P457 = pAAV_RC_1.2SDMSalI_VP2-ko clone 1 c= 308,46 ng/µl
- P458 = pAAV_RC_1.2SDMSalI_VP2-ko clone 2 c= 342,85 ng/µl
- P459 = pAAV_RC_CapIns_prepSDM_VP2-ko clone 1 c= 386,43 ng/µl
- P460 = pAAV_RC_CapIns_prepSDM_VP2-ko clone 2 c= 327,22 ng/µl
- P461 = pAAV_RC_CapIns_noRep_VP2-ko clone 1 c= 373,94 ng/µl
- P462 = pAAV_RC_CapIns_noRep_VP2-ko clone 2 c= 347,39 ng/µl
- P463 = pAAV_RC_RepCapIns_SDMKpnI_VP2-ko clone 1 c= 372,55 ng/µl
- P464 = pAAV_RC_RepCapIns_SDMKpnI_VP2-ko clone 2 c= 348,33 ng/µl
- P465 = pSB1C3_SEG c= 208,32 ng/µl
- P466 = pSB1C3_mGMK c= 224,62 ng/µl
- P470 = pCerulean_VP1up_NLS_Zegfr:1907_VP2/3 clone 1 c= 53,04 ng/µl
- P471 = pCerulean_VP1up_NLS_Zegfr:1907_VP2/3 clone 2 c= 114,24 ng/µl
- P472 = pCerulean_VP1up_NLS_Zegfr:1907_VP2/3-Capins clone 1 c= 170,16 ng/µl
- P473 = pCerulean_VP1up_NLS_Zegfr:1907_VP2/3-Capins clone 2 c= 145,98 ng/µl
- P474 = pCerulean_VP1up_NLS_6xHis_VP2/3 clone 1 c= 104,29 ng/µl
- P475 = pCerulean_VP1up_NLS_6xHis_VP2/3 clone 2 c= 147,86 ng/µl
- P476 = pCerulean_VP1up_NLS_6xHis_VP2/3-Capins clone 1 c= 97,33 ng/µl
- P477 = pCerulean_VP1up_NLS_6xHis_VP2/3-Capins clone 2 c= 97,94 ng/µl
- P478 = pCerulean_VP1up_NLS_mVenus_VP2/3 clone 1 c= 133,61 ng/µl
- P479 = pCerulean_VP1up_NLS_mVenus_VP2/3 clone 2 c= 107,60 ng/µl
- P480 = pCerulean_VP1up_NLS_mVenus_VP2/3-Capins clone 1 c= 45,80 ng/µl
- P481 = pCerulean_VP1up_NLS_mVenus_VP2/3-Capins clone 2 c= 115,16 ng/µl
Test digestion:
No test digestion was performed for Plasmid numbers P449 to P466. P449 to P464 were obtained from a SDM to knock-out VP1 or VP2. There is no possibility to verify this from a test digestion. P465 and P466 were obtained from glycerol stock and therefore no test digestion is necessary.
The other constructs were digested using EcoRI and SpeI for 40 minutes at 37°C.
| components | P470-P481 |
| DNA | 2 |
| Buffer 1 (10x) | 1 |
| BSA (10x) | 1 |
| EcoRI | 0,5 |
| SpeI | 0,5 |
| H2O | 5 |
| Total volume | 10 |
Preparation of gel:
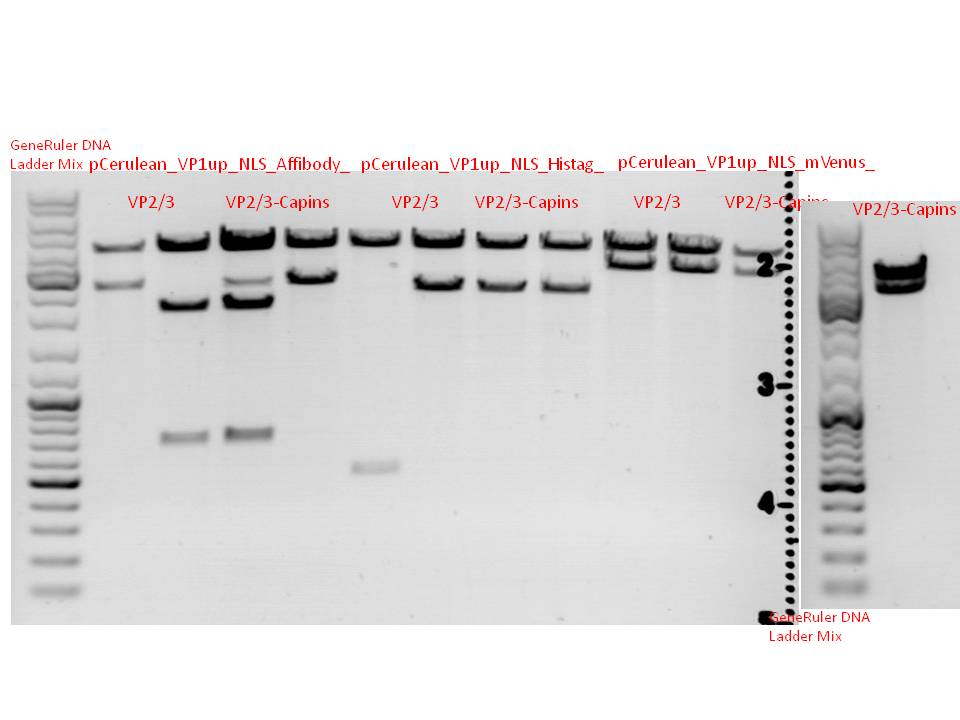
Comment: it is still not clear whether or not the correct insert was cloned into pCerulean. Therefore, P475 and P473 will be sent for sequencing tomorrow using pEGFP_C2-RP as primer to verify VP2/3 insertion.
Sent for sequencing:
Primer used:
2800 reverse (O38)
- P449
- P451
- P453
- P455
- P457
- P459
- P461
- P463
121. labday 15.09.2010
New ViralBrick cloning attempt
Investigator: Achim
Yet another attempt at hybridizing and ligating the ViralBrick oligos into the pSB1C3_BLA. Changes:
- Dephosphorylation of digested vector to prevent religation
- New Klenow Fragment from Fermentas
- Two parallel ligation approaches:
- Quickligation for 10 min
- T4 Ligation over night
Following steps were carried out:
- Hybridisatzion of oligos
- 11:453 Z34C: 143 & 144
- 12:587 Z34C: 145 & 146
- 13:587 KO Z34C: 147 & 148
- 14:587 KO Z34C: 149 & 150
Condtitions as before:
- 2 µg DNA, (3,3 µl of each oligo)
- 2 µl Klenow buffer
- 0,1 µl dNTPs (1µl of 1:10 dilution of our aliquots)
- 9,8 µl H2O
Total volume: 20 µl
Hybridisation was carried out on the gradient cycler using the program "tkfill"
0,5 µl Klenow Fragment was added after the temperature reached 37°C. Klenow reaction was carried out for 1 h in the 37°C room.
Afterwards, the samples were purified with the Quiagen PCR Purification kit. Elution in 50 µl H2O. Nanodrop concentrations:
- 11: 47.08 ng/µl
- 12: 28.50 ng/µl
- 13: 37.69 ng/µl
- 14: 42.98 ng/µl
1 µl of SspI & SalI for sample 11/ BamHI & PvuII for samples 12,13,14 as well as 5,8 µl buffer 4 for each sample were added to the purified samples. Digestion for 3 h at 37 °C
The vector backbone pSB1C3_VCK_BLA (P320)was also digested with BamHI/PvuII and SspI/SalI for 3 h. Afterwards,the cut vector fragment was dephosphorylated with Antarctic Phosphatase for 1h at 37°C.
The oligos and the vector fragments were mixed with loading dye and run over a preparative gel (1%):
Fragments were cut out, followed by gel extraction. Two ligations were prepared: T4-Ligation and Quickligation.
Quickligation: 10 min, followed by trafo. Plates were incubated overnight.
T4-Ligation: Overnight
Cloning VP2/3 approaches into pSB1C3_VCK vector
Investigator: Bea
Comment:Since the constructs VP2/3 and VP2/3_ins_cap were cloned in the plasmid pSB1C3 they need to recloned into the vector pSB1C3_VCK which have two deleted restrictions sites (PvuII ans SspI) . These restriction sites have to be single cutting enzymes in this case because we want to subclone the HSPG-knockout "motif" into the cap gene in order to obtain constructs for the N-terminal targeting approach. The recombinant virus particles basically then do not show the natural tropism for its primary receptor.
Digestion of the constructs:
- P440 = pSB1C3_VP2/3 c=290 ng/µL
- P440 = pSB1C3_VP2/3_ins_cap c=314ng/µL
- P309 = pSB1C3_VCK_BLA c=408ng/µL
| Components | vP440 /µL | vP444/µL | vP309 /µL |
| DNA | 5,5 | 5,5 | 2,5 |
| BSA (10x) | 4 | 4 | 3 |
| Buffer no. 4 (10x) | 4 | 4 | 3 |
| EcoRI | 1 | 1 | 1 |
| AgeI | 1 | 1 | 1 |
| MISC | SacI: 1 | MscI: 1 | - |
| H2O | 23,5 | 23,5 | 19,5 |
| Total volume | 40 | 40 | 30 |
The expected fragments of the digested constructs were: The underlined fragments represent the fragments which correspond to the desired construct which can be used for ligation.
- P440: 1950, 1158, 908bp
- P444: 1950, 1347, 715bp
- P309: 2067, 894bp
Loading plan:
M P440 P444 P309
Results:
After gel extraction has been performed, the ligation was carried out.
Ligation:
- v=P444 =5,5µL
- v=P309 =2,5µL
The ligation mix was transformed into XL1-Blue cells and plated on agar plates containing chloramphenicol.
Next steps:
Picking clones and perform Mini-Prep. After the Mini-Preps have been performed these constructs can be used for inserting the HSPG knockout motif.
Colony PCR of CD colonies
Investigator: Kira
Comments: Since the last approach with the CD did not work, we performed a colony PCR in order to detect the positive clones which should contain the CD gene. We used the two SDM primers, originally used for deleting the iGEM restriction sites in the CD gene, which should only bind to the CD sequence.
15 colonies were picked from the plate and dispersed first into the PCR tube and then into DYT media as a backup in case of successful PCR reaction.
| components | pro sample |
| 10x Taq Buffer | 1 |
| MgCl (50mM) | 0,1 |
| dNTPs | 0,2 |
| Primer 1 | 0,2 |
| Primer 2 | 0,2 |
| Taq Polymerase | 0,05 |
| H2O | 8,25 |
| Total volume | 10 |
PCR program:
| Cycles | Temperature | Time |
| 95°C | 6 min | |
| 30x | 95°C | 25 sec |
| 63°C | 30 sec | |
| 68°C | 1 min 6 sec | |
| 1x | 68°C | 5 min |
| Hold 4°C |
pSB1C3_RFC25_CFP was used as a control sample
1% agarose gel
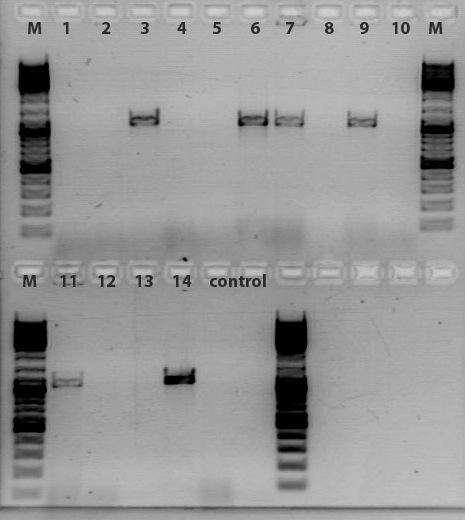
According to the gel, clone 3, 6, 7, 9, 11, 14 contain CD gene. These samples will be incubated over-night @ 37C.
Comment added on 17.09.2010 : We (Bea, Kira, Stefan and Tobi) wondered why sequencing of one of the positive clones of the colony PCR revealed that the construct did not contain the expected CD gene but CFP instead. Today, we realized that the colony PCR does not reveal only positive constructs which contain the cytosine deaminase. Problem is that we received the CD gene originating from the COD-BA gene of E. coli. Since we performed a colony PCR with XL1-B E. coli cells the used primers will bind to the genomic CD-gene aswell which could explain the positive clones in the gel, the negative results of seuqencing and another test digest of hte sequenced clone.
Next steps are that we are performing the colony PCR again with new primers which means that we are using the iGEM primers which should specifically bind to the constructs which contain the iGEM prefix and suffix add-ons. Since CFP does contain the iGEM restriction sites aswell, but differ in the size of the expected fragment (CD ~1100bp vs. CFP ~700bp) the positive clones should be detectable by the size length.
Cloning of pSB1C3_Affibody_VP2/3
Investigator: Jessica
Comment: Cloning of Affibody and VP2/3 without linker to test if it works without linker
Digestion:
| components | pSB1C3_ZEGFR:1907 clone 2 (P268) | pSB1C3_VP2/3 clone 1 (P440) |
| DNA | 14,35 | 5,2 |
| BSA | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| XbaI | 1 | 1 |
| NgoMIV | - | 1 |
| AgeI | 1 | - |
| H2O | - | 8,8 |
| Total volume | 20,35 | 20 |
Comment:Digestion was performed 1,5 hours at 37 °C
Gel:
0,5 g Agarose,50 ml TAE (1%), 5 µl Gelred , at Volt
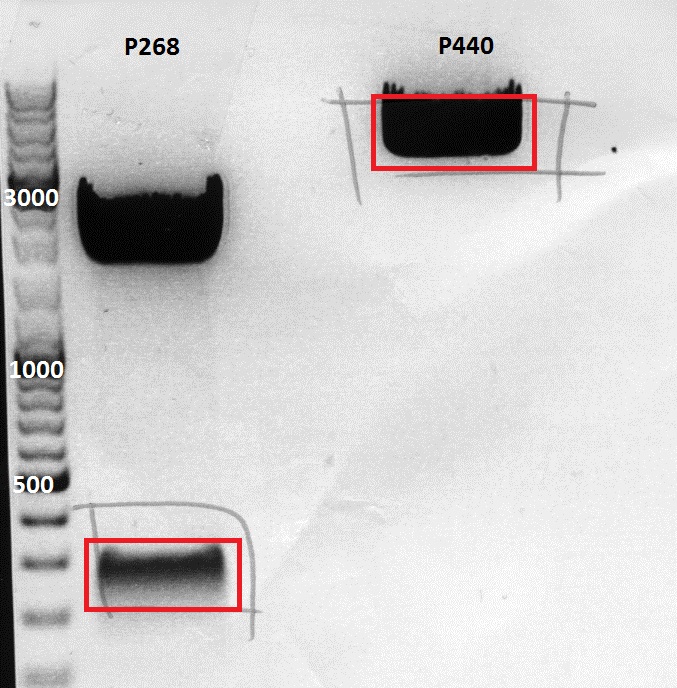
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- pSB1C3_ZEGFR:1907 clone 2 (P268): c= 2,6ng/µl
- pSB1C3_VP2/3 clone 1 (P440: c= 6,7ng/µl
Ligation:
The Ligation was performed as following:
- Vector Volume: 5,83µl
- Insert Volume: 2,17µl
- 1 µl T4Ligase buffer (2x)
- 8 µl (Vector + Insert) mix
- 1 µl T4Ligase
Incubating for 45 minutes.
Transformation:
Trafo was performed according to the standard protocol (XL1b). The cells were plated on a agar plate with Cm.
PCR of GFP_RFC25 and cloning into pSB1C3_6XHis and pSB1C3_CFP
Investigator: Anissa
Comment:GFP will be cloned into pSB1C3 for biobrick production and upstream of his-tag into into pSB1C3_his for further use
Preparations:
A 1:1000 dilution of the plasmid P416 (pSB1A3_EGFP_HQ clone1). GFP will be extracted and cloned into pSB1C3 iGEM standard plasmid with the RFC25 standard.
PCR protocol:
| Ingredients | volume |
| 5X Phusion HF buffer | 10 µl |
| 10 mM dNTP mix | 1µl |
| forward primer: O186 | 2,5 µl |
| reverse primer: O187 | 2,5 µl |
| DNA Template | 5 µl |
| DMSO | 0 µl |
| Phusion Polymerase | 0,5 µl |
| H2O | 28,5 µl |
| Total volume | 50 µl |
The following PCR programm was used. Idea is that in the first 8 cycles the primer can only bind to the template added in to the reaction mix. The following 17 cycles afterwards allow the annealing of the complete primer containing prefix and suffix with its restriciton sites.
| Cycles | Temperature | Time |
| 98°C | 1 | |
| 8x | 98°C | 15" |
| 54°C | 25" | |
| 72°C | 24" | |
| 17x | 98°C | 15" |
| 64°C | 25" | |
| 72°C | 7" | |
| 1x | 72°C | 5' |
| Hold 4°C |
Digestion of the vectors:
| components | pSB1C3_CFP (P51.2) | pSB1C3_6XHis (P300) |
| DNA | 6,6 | 5,6 |
| BSA | 1,5 | 1,5 |
| Buffer 4 (10x) | 1,5 | 1,5 |
| XbaI | 1 | 1 |
| NgoMIV | - | 1 |
| AgeI | 1 | - |
| H2O | 3,4 | 4,4 |
| Total volume | 15 | 15 |
Comment:Digestion was performed 1,5 hours at 37 °C
Gel:
0,5 g Agarose,50 ml TAE (1%), 3 µl Gelred , at 110 Volt
Comment:The gel showed for the sample p300 no clear band. Perhaps the enzymes did not cut right. The gel-ex-sample of p300 was seperated into an "upper" and a "lower band" sample.
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- pSB1C3 (P51.2): c= 3,95 ng/µl
- pSB1C3_6Xhis_upper (P300): c= 8,46 ng/µl
- pSB1C3_6Xhis_lower (P300): c= 2,10 ng/µl
- PCR GFP_RFC25: c= 6 ng/µl
Digestion of the PCR-product:
| components | GFP_RFC25 |
| DNA | 30 |
| BSA | 4 |
| Buffer 4 (10x) | 4 |
| XbaI | 1 |
| AgeI | 1 |
| H2O | - |
| Total volume | 40 |
Comment:Digestion was performed 1,5 hours at 37 °C
Purification of the PCR-product:
Was performed accoding to the standard-protocol
Ligation:
The Ligation was performed as following:
- P51.2 Volume: 5,83µl
- Insert Volume: 2,17µl
- P300_upper Volume: 5,83µl
- Insert Volume: 2,17µl
- P300_lower Volume: 5,83µl
- Insert Volume: 2,17µl
- 1 µl T4Ligase buffer (2x)
- 8 µl (Vector + Insert) mix
- 1 µl T4Ligase
Incubating for 30 minutes.
Transformation:
Trafo was performed according to the standard protocol (XL1b). The cells were plated on a agar plates .
PCR for biobrick production: Cloning RepCap into pSB1C3
Investigator: Stefan
Comment:Goal of the experiment was to bring the different RepCap constructs into the pSB1C3 plasmid containing the deletions of the restriction sites interfering with the loop insertions.
Preparations:
Plasmids used:
- pAAV_RC_1.2SDMSalI (P158)
- pAAV_RC_CapIns_prepSDM (P396)
- pAAV_RC_CapIns_noRep (P413)
- pAAV_RC_RepCapIns_SDMKpnI (P432)
A 1:1000 dilution of each of the plasmids was prepared.
PCR protocol:
| Ingredients | P158 (v/µl) | P396 (v/µl) | P413 (v/µl) | P432 (v/µl) |
| 5X Phusion HF buffer | 10 | 10 | 10 | 10 |
| 10 mM dNTP mix | 1 | 1 | 1 | 1 |
| forward primer: O93 | 2,5 | 2,5 | 2,5 | 2,5 |
| reverse primer: O120 | 2,5 | 2,5 | 2,5 | 2,5 |
| DNA Template | 2,5 | 2 | 3,2 | 3 |
| DMSO | - | - | - | - |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 31 | 31,5 | 30,3 | 30,5 |
| Total volume | 50 | 50 | 50 | 50 |
PCR program:
Comment:For all approaches the following programm was used:
| Cycles | Temperature /°C | Time /s |
| 1 | 98 | 60 |
| 2 (8x step 2-4) | 98 | 15 |
| 3 | 59 | 25 |
| 4 | 72 | 70 |
| 5 (17x step 5-6) | 98 | 15 |
| 6 | 72 | 80 |
| 71x | 72 | 300 |
| Hold 4°C |
Digestion of the vector:
| components | pSB1C3_VCK_Bla (P320) (v/µl) |
| DNA | 6,1 |
| BSA | 2 |
| Buffer 4 (10x) | 2 |
| XbaI | 1 |
| AgeI | 1 |
| H2O | 7,9 |
| Total volume | 20 |
Comment:Digestion was performed 2 hours at 37 °C.
Gels:
For PCR products:
0,4 g Agarose,50 ml TAE (0,8%), 3 µl Gelred , at 110 Volt; run for ~40 minutes
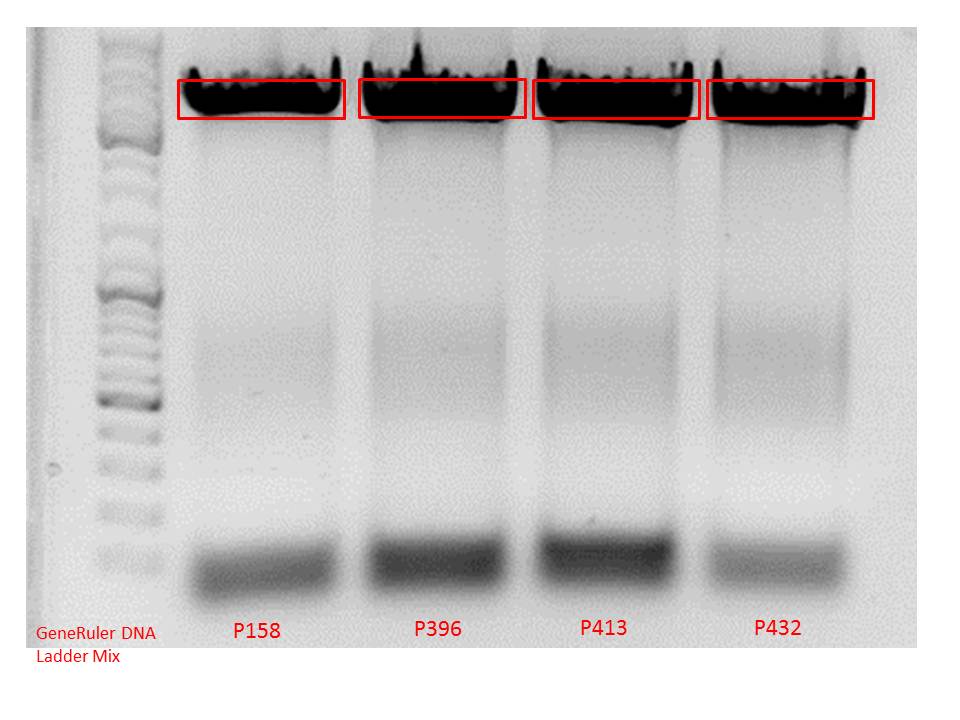
For vector:
0,5 g Agarose,50 ml TAE (1%), 3 µl Gelred , at 110 Volt; run for ~50 minutes
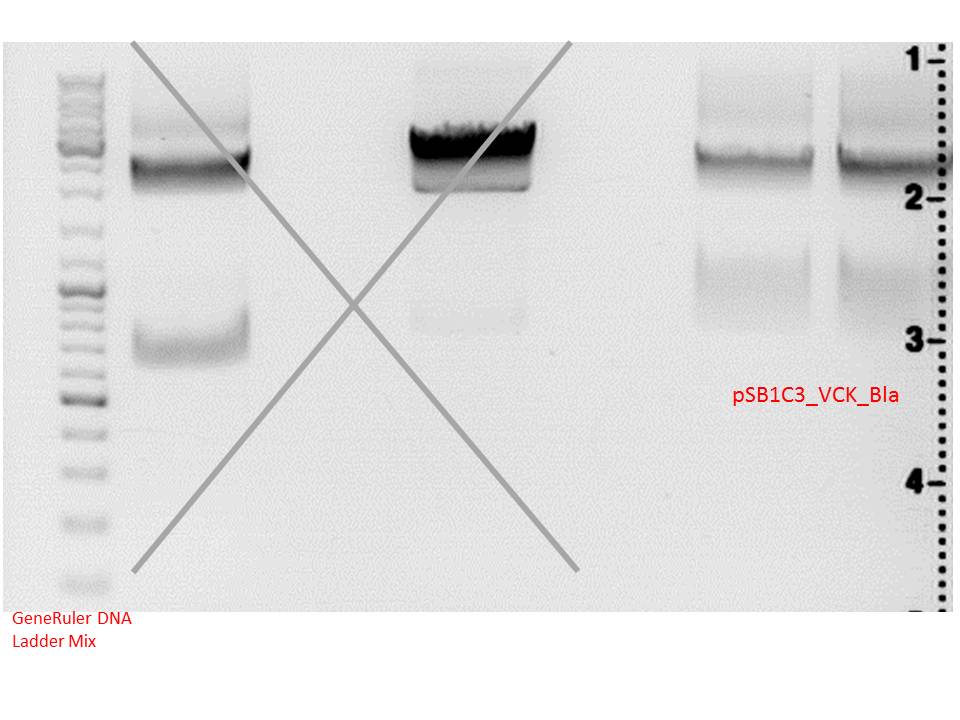
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the vector:
- pSB1C3 (P320): c= 11,97 ng/µl
Digestion of the PCR-products:
Comment:Digestion was performed for all PCR products alike.
| components | PCR products (v/µl) |
| DNA | 30 |
| BSA | 4 |
| Buffer 4 (10x) | 4 |
| XbaI | 1 |
| AgeI | 1 |
| H2O | - |
| Total volume | 40 |
Comment:Digestion was performed 2 hours at 37 °C.
PCR purification:
Purification was performed accoding to the standard protocol. The following concentrations were measured:
- PCR of P158: c= 7,68 ng/µl
- PCR of P396: c= 17,65 ng/µl
- PCR of P413: c= 24,17 ng/µl
- PCR of P432: c= 18,23 ng/µl
Ligation:
1 µl T4 Ligase buffer (2x)
8 µl DNA mix
1 µl T4 Ligase
Incubating for 45 minutes at room temperature.
The Ligation was performed using the following amounts of DNA:
- Vector Volume: 1,98 µl
- P158 Volume: 6,02 µl
- Vector Volume: 3,45 µl
- P396 Volume: 4,55 µl
- Vector Volume: 4,07 µl
- P413 Volume: 3,93 µl
- Vector Volume: 3,51 µl
- P413 Volume: 4,49 µl
Transformation:
Trafo was performed according to the standard protocol using XL1b cells. The cells were plated on a agar plates containing chlorampenicol.
Mini-Preps
Investigator: Hanna
Mini-Preps were performed according to standard protocol:
pSB1C3_ITR_phTERT_b-globin__mGMK clone 1 (P483)
pSB1C3_ITR_phTERT_b-globin__mGMK clone 2 (P484)
pSB1C3_lITR_phTERT_b-globin__mVenus clone 1 (P485)
pSB1C3_lITR_phTERT_b-globin__mVenus clone 2 (P486)
pSB1C3_lITR_CMV_b-globin__mGMK clone 1 (P487)
pSB1C3_lITR_CMV_b-globin__mGMK clone 2 (P488)
pSB1C3_VP123 (aus pAAV_RC_CapIns_noRep P413) clone 1 (P489)
pSB1C3_VP123 (aus pAAV_RC_CapIns_noRep P413) clone 2 (P490)
pSB1C3_VP123 (aus pAAV_RC_1.2 SDM SalI P158) clone 1 (P491)
pSB1C3_VP123 aus (pAAV_RC_1.2 SDM SalI P158) clone 2 (P492)
Colony PCR of pCerulean_CFP_MiddleLinker, pCerulean_6xHis_MiddleLinker and pCerulean_"Linker"_Zegfr:1907 constructs (Linker library)
Investigator: Hanna
Comment: Because we found out that there's also a SacI restriction site in the VP2/3 sequence - due to the PstI (4073) mutagenesis - the first try of cloning VP2/3 to the C-terminus of the Zegfr:1907, CFP and His-Tag constructs was discarded.
Fortunately Anissa already repeated this experiment yesterday. Because we're not sure whether cloning was successful (one plate was without any colonies and has to be done one more time), we decided to perform a colony PCR before continuation with (at least 22!) mini-preps. We chose primer which bind in the VP2/3 region and therefore amplify a VP2/3 fragment - given, that VP2/3 was successfully cloned downstream of the linker. One further advantage is, that we can also save the test digestions.
A Mastermix was prepared for 43 samples:
- 5x Crimson Taq Mg-free reaction buffer: 215 µL
- 10 mM dNTP: 21.5 µL
- 10 µM Forward primer (Primer Cap 3500 for): 21.5 µL
- 10 µM Reverse Primer (Primer Cap 4200 rev): 21.5 µL
- Crimson Taq DNA Polymerase: 5.375 L
- Nuclease-Free water: 790.125 µL
Total: 1075 µL -> 25 µL reactions
2-4 colonies were picked with a tooth pick from each plate (11). 25 µL master mix and 5 ml LB + Kanamycin were inocculated.
PCR program:
According to standard protocol provided by Gerrit
- 95°C, 30
- 95°C 25
- 50.5°C, 40
- 68°C, 60 --> repeat 2-4 27 times
- 68°C, 300
- 4°C, hold
Tomorrow morning the PCR products will be tested on an analytic gel. If the samples will be positive, they will be mini-preped - if not cloning will be done again.
Tod do: Cloning of VP2/3 into pCerulean_CFP_MiddleLinker.
Sequencing analysis of VP1-ko and VP2-ko in severeal pAAV vectors
Investigator: Stefan
Primer used (for all plasmids): 2800 rev (O38)
Sequenced plasmid:
- pAAV_RC_1.2SDMSalI_VP1-ko clone 1 (P449)
- pAAV_RC_CapIns_prepSDM_VP1-koclone 1 (P451)
- pAAV_RC_CapIns_noRep_VP1-ko clone 1 (P453)
- pAAV_RC_RepCapIns_SDMKpnI_VP1-ko clone 1 (P455)
- pAAV_RC_1.2SDMSalI_VP2-ko clone 1 (P457)
- pAAV_RC_CapIns_prepSDM_VP2-ko clone 1 (P459)
- pAAV_RC_CapIns_noRep_VP2-ko clone 1 (P461)
- pAAV_RC_RepCapIns_SDMKpnI_VP2-ko clone 1 (P463)
Comment:All sequences showed that the site directed mutagenesis worked out for all constructs. All sequencing results revealed problems in the area shown below:
122. labday 16.09.2010
Trafo of T4 ligation, Results of Quickligation
Investigator: Achim
No clones on the Quickligation plates. Trafo of T4 Ligation was carried out in XL1B cells. Results will be evaluated tomorrow.
Mini Prep and test digestion of several constructs
Investigator: Anissa, Bea
Comment: Several test digestions have been performed for verifying the cloned fragment into the plasmid backbone. The constructs for reassembling the vector plasmid (pSB1C3_leftITR_PROMOTER_betaglobin_GOI) were digested with EcoRI and NgomIV, the SV40 construct was digested with EcoRI and PstI.
Test digestion of the constructs:
- pSB1C3_leftITR_phTERT_betaglobin_mVenus (P485-486)
- pSB1C3_leftITR_phTERT_betaglobin_mGMK (P483-484)
- pSB1C3_leftITR_pCMV_betaglobin_mGMK (P487-488)
- pSB1C3_SV40 (P493-494)
Expected size fragments are:
- (P485-486): 1150bp, 2780bp
- (P483-484): 1150bp, 2650bp
- (P487-488): 1300bp, 2600bp
- (P493-494): 270bp, 2030bp
Results: Test digestion of the constructs looks well, except for the construct 494 which is one clone of the construct pSB1C3_SV40. Since the other clone is fine, the construct was discarded. The control K437 is the plasmid pSB1C3_leftITR_hTERT_beta-globin without the assembled GOI (mGMK and mVenus repsectively). The GOI´s are in the RFC25 standard therefore leading to scar with a additional NgoMIV after fusing it to the before mentioned construct which is in the RFC10 standard. Therefore, test digestion was carried out with NgoMIV and EcoRI leading to fragments which can be separated from the backbone in a 1% agarose gel. The control vector should only be linearized because it does not contain any NgomIV rstriction site.
Test digestion of pSB1C3_VP123
Investigator: Jessica
| Samples | Plasmid | Clone | Concentration [ng/µl] |
| P489 | pSB1C3_VP123 (aus pAAV_RC_CapIns_noRep P413) | 1 | 350 |
| P490 | pSB1C3_VP123 (aus pAAV_RC_CapIns_noRep P413) | 2 | 395,5 |
| P491 | pSB1C3_VP123 (aus pAAV_RC_1.2 SDM SalI P158) | 1 | 403 |
| P492 | pSB1C3_VP123 (aus pAAV_RC_1.2 SDM SalI P158) | 2 | 411 |
Test digestion:
| components | Volume for each sample /µl |
| DNA | 2,6/2,3/2,3/2,2 |
| BSA (10x) | 1 |
| Buffer 4 (10x) | 1 |
| Enzyme NgoMIV | 0,5 |
| Enzyme SapI | 0,5 |
| H2O | 4,4/4,7/4,7/4,8 |
| Total volume /µl | 10 |
Preparation of gel:
- Incubation time: 1,5 h, Incubation temperature: 37°
- 0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at Volt, running time: minutes
- Expected fragment sizes:
3244 bp
767 bp
Comment: Sample P489 was sent for sequencing with primer VR2. Sequencing result looks good, VP123 is in
Continuation of preparation of competent E.coli
Investigator: Jessica
preparation of competent BL21 was finished according to the standard protocol
- aliquots of 60µl (to use for 1 trafo) are stored in -80°C freezer
don't use before results of testtrafo is writen down
testtrafo was made with pUC and Amp
Mini prep of CD-clones, which were identified by colony pcr
Investigator: Kira
c(clone1) = 160,29 ng/ul
c(clone2) = 167,98 ng/ul
c(cloone3) = 181, 51 ng/ul
clone 3 was sent for sequencing
Repetition: PCR for biobrick production of P40 promotor
Investigator: Stefan
Comment: During trouble-shooting we realized that the reverse primer P40_rev (O153) was designed forward and therefore does not work. A new primer will have to be designed and ordered to repeat the PCR.
Results of Colony PCR
Investigator: Hanna
Comment: In order to find out, whether the VP2/3 or the VP2/3_Capins fragment was successfully cloned into pCerulean_Zegfr:1907_"Linker" (--> Linker library), pCerulean_6xHis_MiddleLinker and pCerulean_CFP_MiddleLinker, a colony PCR was performed yesterday. From each plate 2-4 clones were picked, so that we got 41 samples (including one positive control = pAAV_RC). With VP_3500_for and VP_4200_rev primer a 880 fragment should be amplified of each colony, if cloning functioned.
Samples were loaded onto a 1% agarose gel:
- 1. pCerulean_Zegfr:1907_ShortLinker_VP2/3
- 2. pCerulean_Zegfr:1907_MiddleLinker_VP2/3_Capins
- 3. pCerulean_Zegfr:1907_LongLinker_VP2/3
- 4. pCerulean_6xHis_MiddleLinker_VP2/3
- 5. pCerulean_Zegfr:1907_SEG_VP2/3
- 6. pCerulean_6xHis_MiddleLinker_VP2/3_Capins
- 7. pCerulean_CFP_MiddleLinker_VP2/3_Capins
- 8. pCerulean_Zegfr:1907_ShortLinker_VP2/3_Capins
- 9. pCerulean_ Zegfr:1907_SEG_VP2/3_Capins
- 10. pCerulean_Zegfr:1907_LongLinker_VP2/3_Capins
- 11. pCerulean_Zegfr:1907_MiddleLinker_VP2/3
As can be seen in the gel picture, it seemed that just clone 9.3, 10.2, 11.1 and 11.2 were VP2/3 positive. Curiously the positive control (K) was negative.
These 4 clones were mini-preped by Achim and sent for sequencing to GATC: primer = GATC_std_CMV-F.
Because I found out that some things went wrong during VP cloning due to false geneious annotations and theoretical cloning approaches, all pAAV_RC constructs (with/without Cap or Rep-insert, 4-fold mutated, PvuII insertion, KpnI-backmutation,...) were theoretically prepared again (see geneious folder: "pAAV_RC constructs (DO NOT CHANGE!!!)").
In addition to that the theoretical cloning of the final N-terminal fusion approaches to VP2/3 were performed once again with these correct sequences. With the help of Sven cloning of the not yet positive approaches were repeated (The ones which were negative in colony PCR).
To do (tomorrow): Pciking clones, analyze sequencing. Check and plan HSPG-KO approaches and Zegfr:1907
Practical Cloning:
- Vector: pCerulean_CFP_MiddleLinker (P407), pCerulean_Zegfr:1907_MiddleLinker (P408), pCerulean_Zegfr:1907_SEG (P409), pCerulean_Zegfr:1907_ShortLinker (P410), pCerulean_Zegfr:1907_LongLinker (P412), pCerulean_6xHis_MiddleLinker (P374)
- Insert: pSB1C3_VP2/3 (P440), pSB1C3_VP2/3_ins_cap (P444)
- new vector name: pCerulean_CFP_MiddleLinker_VP2/3, pCerulean_Zegfr:1907_MiddleLinker_VP2/3, pCerulean_Zegfr:1907_SEG_VP2/3, pCerulean_Zegfr:1907_ShortLinker_VP2/3, pCerulean_Zegfr:1907_LongLinker_VP2/3, pCerulean_6xHis_MiddleLinker_VP2/3 AND pCerulean_CFP_MiddleLinker_VP2/3-insCap, pCerulean_Zegfr:1907_MiddleLinker_VP2/3-insCap, pCerulean_Zegfr:1907_SEG_VP2/3-insCap, pCerulean_Zegfr:1907_ShortLinker_VP2/3-insCap, pCerulean_Zegfr:1907_LongLinker_VP2/3-insCap, pCerulean_6xHis_MiddleLinker_VP2/3-insCap
- buffer used:4
Comment Because the VP2/3 frament has approximately the same size as the backbone pSB1C3 it may be difficult to separate them on the agarose gel. Therefore a single cutter enzyme (MscI) was chosen to digests the backbone. This enzyme also works in buffer 4 + BSA and needs to be incubated at 37°C.
- Vector: AgeI, SpeI
- Insert: NgoMIV, SpeI, SacI
Digestion
- Incubation: 2 h
122. labday 17.09.2010
Sequencing analysis of pSB1C3_VP123
Investigator: Bea
Comment: The constrcuct was created by performing a PCR with the pAAV_RC_inscap construct. The sequence analysis reveals that the suffix looks well. Additionally the mutation from TAA to TGA can be seen.
ViralBricks: Picking clones from T4 ligation
Investigator: Achim
Sequencing results N-terminal fusion - part 1
Investigator: Hanna
Comment: Colony PCR delivered that 4 clones contained the VP2/3 or VP2/3_Capins sequence. These clones were sent for sequencing:
1. pCerulean_Zegfr:1907_LongLinker__VP2/3_Capins:
Sequencing looked well: VP2/3_Capins was successfully fused to the C-terminus of the Long-Linker. The additional basepair at the beginning of the Affibody was misinterpreted (as one can see in the chromatogram) by Geneious.
2. pCerulean_Zegfr:1907_MiddleLinker__VP2/3 clone 1:
Sequencing looked well: VP2/3 was successfully fused to the C-terminus of the Middle-Linker. The additional basepair at the beginning of the Affibody was again misinterpreted (as one can see in the chromatogram) by Geneious.
3. pCerulean_Zegfr:1907_MiddleLinker__VP2/3 clone 2:
Sequencing looked well: VP2/3 was successfully fused to the C-terminus of the Middle-Linker. The additional basepair at the beginning of the Affibody was again misinterpreted (as one can see in the chromatogram) by Geneious.
4. pCerulean_Zegfr:1907_SEG__VP2/3_Capins:
Sequencing looked well: VP2/3_Capins was successfully fused to the C-terminus of the SEG-Linker. The additional basepair at the beginning of the Affibody was again misinterpreted (as one can see in the chromatogram) by Geneious.
Mini-Prep and test digestion of several constructs
Investigator: Stefan
Glycerol stocks were prepared:
- B430 = pSB1C3_001_RC_4fold mutated clone 1
- B431 = pSB1C3_001_RC_4fold mutated clone 2
- B432 = psB1C3_001_RC_InsRepCap clone 1
- B433 = psB1C3_001_RC_InsRepCap clone 2
- B434 = pSB1C3_001_RC_InsCap clone 1
- B435 = pSB1C3_001_RC_InsCap clone 2
- B436 = pSB1C3_001_RC_InsRepCap_KpnIback clone 1
- B437 = pSB1C3_001_RC_InsRepCap_KpnIback clone 2
- B438 = pSB1C3_001_VP2/3insCap clone 1
- B439 = pSB1C3_001_VP2/3insCap clone 2
- B440 = pSB1C3_ZEGFR:1907_VP2/3 clone 1
- B441 = pSB1C3_ZEGFR:1907_VP2/3 clone 2
- B442 = pSB1C3_GFP_RFC25_upper band clone 1
- B443 = pSB1C3_GFP_RFC25_upper band clone 2
- B444 = pSB1C3_GFP_RFC25_lower band clone 1
- B445 = pSB1C3_GFP_RFC25_lower band clone 2
- B446 = pSB1C3_GFP_RFC25 clone 1
- B447 = pSB1C3_GFP_RFC25 clone 2
Mini-Prep was performed according to the standard protocol
- P506 pSB1C3_001_RC_4fold mutated clone 1
- P507 pSB1C3_001_RC_4fold mutated clone 2
- P508 psB1C3_001_RC_InsRepCap clone 1
- P509 psB1C3_001_RC_InsRepCap clone 2
- P510 pSB1C3_001_RC_InsCap clone 1
- P511 pSB1C3_001_RC_InsCap clone 2
- P512 pSB1C3_001_RC_InsRepCap_KpnIback clone 1
- P513 pSB1C3_001_RC_InsRepCap_KpnIback clone 2
- P514 pSB1C3_001_VP2/3insCap clone 1
- P515 pSB1C3_001_VP2/3insCap clone 2
- P516 pSB1C3_ZEGFR:1907_VP2/3 clone 1
- P517 pSB1C3_ZEGFR:1907_VP2/3 clone 2
- P518 pSB1C3_GFP_RFC25_upper band clone 1
- P519 pSB1C3_GFP_RFC25_upper band clone 2
- P520 pSB1C3_GFP_RFC25_lower band clone 1
- P521 pSB1C3_GFP_RFC25_lower band clone 2
- P522 pSB1C3_GFP_RFC25 clone 1
- P523 pSB1C3_GFP_RFC25 clone 2
- P524 pSB1C3_VP2/3
- P525 pSB1C3_VP2/3_ins_cap
Test digestion:
Test digestion was performed for plasmids P506, P507, P510 to P513 and for P522 and P523. Test digestion of P514 to P521 will be performed tomorrow. Preps for P524 and P525 were inocculated from glycerol stock and therefor no digestion will be performed.
The constructs were digested using XbaI and AgeI for 40 minutes at 37°C.
| components | P470-P481 | P470-P481 |
| DNA | 2 | 2 |
| Buffer 4 (10x) | 1 | 1 |
| XbaI | 0,4 | 0,4 |
| AgeI | 0,4 | 0,4 |
| H2O | 7,2 | 6,2 |
| Total volume | 10 | 10 |
Preparation of gel:
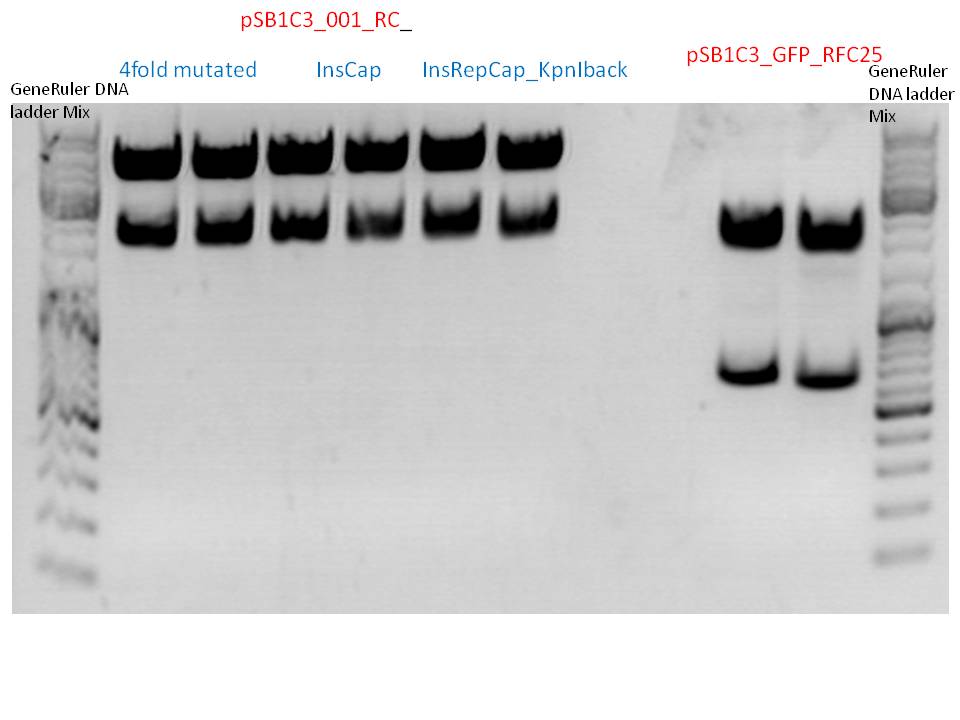
Comment:
Sent for sequencing:
Primer used:
VR2
plasmid no:
P522
tube labeled: SB_1
Sent for sequencing
Investigator: Stefan
pSB1C3_SV40 (P493) was sent for sequencing using VR2 primer. Results have to be checked tomorrow.
Sequencing analysis of VP2/3 insertion into pCerulean_VP1up_NLS_targeting motif
Investigator: Stefan
Comment: Sequences can be found in GATC Watchbox (date 15.10.10) labeled SB_1 (VP2/3 insertion) and SB_2 (VP2/3_insCap insertion).
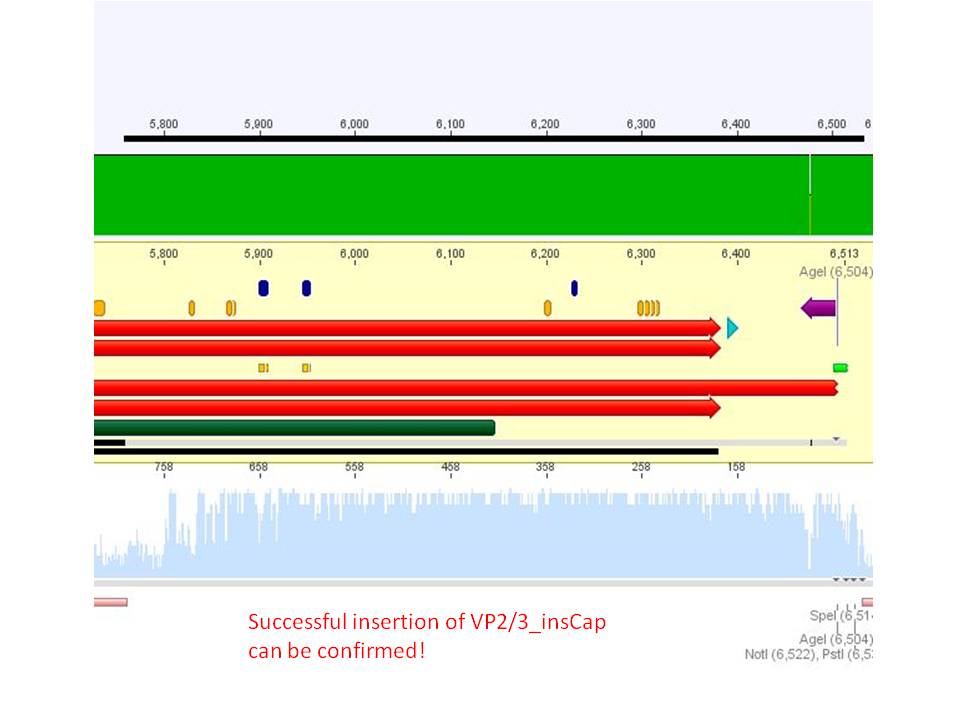
Biobrick production of VP2/3- and VP2/3 cap-Gsat_linker
Motivation: Production of fusion proteins
Investigator: Kira
Digestions of backbones and inserts:
| Components | VP2/3 | VP2/3 cap | Gsat linker |
| DNA | 7 | 7 | 7 |
| BSA (100x) | 0 | 0 | 0 |
| Buffer no. 4 (10x) | 2,5 | 2,5 | 2,5 |
| EcoRI | 0,5 | 0,5 | 0,5 |
| NgoMIV | 0,5 | 0,5 | 0 |
| XbaI | 0 | 0 | 1,25 |
| H2O | 14,5 | 14,5 | 13,75 |
| Total volume | 25 | 25 | 25 |
incubation @ 37 C for approx. 3 h
1% agarose gel
Ligation:
T4 ligase was used. ligation over night @ 18 C
VP2/3-mix 8 ul (3,7 ul vector + 4,3 ul insert)
VP2/3 cap-mix 8 ul (3,7 ul vector + 4,3 ul insert)
123. labday 18.09.2010
Continuation of preparation of competent E.coli
Investigator: Jessica
preparation of competent XL1blue was finished according to the standard protocol
- aliquots of 60µl (to use for 1 trafo) are stored in -80°C freezer
don't use before results of testtrafo is writen down
Test digestion of pSB1C3_ZEGFR:1907_VP2/3
Investigator: Jessica
| Samples | Plasmid | Clone | Concentration [ng/µl] |
| P514 | pSB1C3_001_VP2/3insCap | 1 | 318.9 |
| P515 | pSB1C3_001_VP2/3insCap | 2 | 321.8 |
| P516 | pSB1C3_ZEGFR:1907_VP2/3 | 1 | 191,6 |
| P517 | pSB1C3_ZEGFR:1907_VP2/3 | 2 | 210,8 |
| P518 | pSB1C3_GFP_RFC25_upper band | 1 | 138.3 |
| P519 | pSB1C3_GFP_RFC25_upper band | 2 | 72.3 |
| P520 | pSB1C3_GFP_RFC25_lower band | 1 | 141.6 |
| P521 | pSB1C3_GFP_RFC25_lower band | 2 | 127.5 |
Test digestion:
| components | Volume for each sample P516/517 /µl | Volume for each sample P514/515 /µl | Volume for each sample P518-521 /µl |
| DNA | 2,5 | 3,5 | 1,5 |
| BSA (10x) | 1 | 1 | 1 |
| Buffer 1/4/4 (10x) | 1 | 1 | 1 |
| Enzyme NgoMIV | 0,5 | - | - |
| Enzyme DraIII | 0,5 | - | - |
| Enzyme XbaI | - | - | 0,4 |
| Enzyme AgeI | - | - | 0,4 |
| Enzyme MscI | - | 0,4 | - |
| Enzyme SpeI | - | 0,4 | - |
| H2O | 4,5 | 3,7 | 5,7 |
| Total volume /µl | 10 | 10 | 10 |
Preparation of gel:
- Incubation time: h, Incubation temperature: 37°
- 0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at Volt, running time: minutes
- Expected fragment sizes:
P516/517: 3540bp / 651bp
P514/515: 1300bp / 2700bp
P518-521: 2000bp / 700bp
Comment: Sample was sent for sequencing with primer .
Mini-Prep of pSB1C3_SV40 clone 1 (B420)
Investigator: Jessica
was done because of the low concentration of the plasmid P493
- P526: pSB1C3_SV40 clone 1 (=B420) c= 176,0 ng/µl
New aliquots (60µl) of BL21 can be used ! (Jessica)
 "
"