Team:Freiburg Bioware/NoteBook/Labjournal/October
From 2010.igem.org
- March (labday 1)
- April (labday 2 - 5)
- May (labday 6 - 17)
- June (labday 18 - 45)
- July (labday 46 - 75)
- August part 1 (labday 76 - 92)
- August part 2 (labday 93 - 106)
- September part 1 (labday 107 - 123)
- September part 2 (labday 124 - 135)
- October part 1 (labday 136 - 149 )
- October part 2 (labday 150 - 166 )
- November (labday 167 - 170 )
- Cellculture
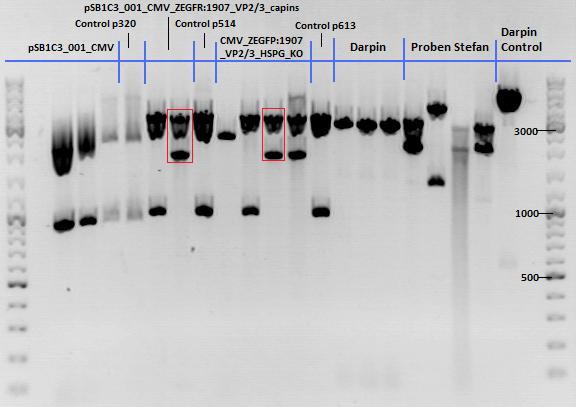
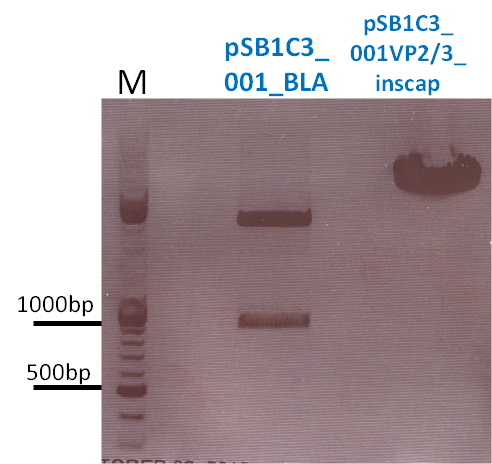
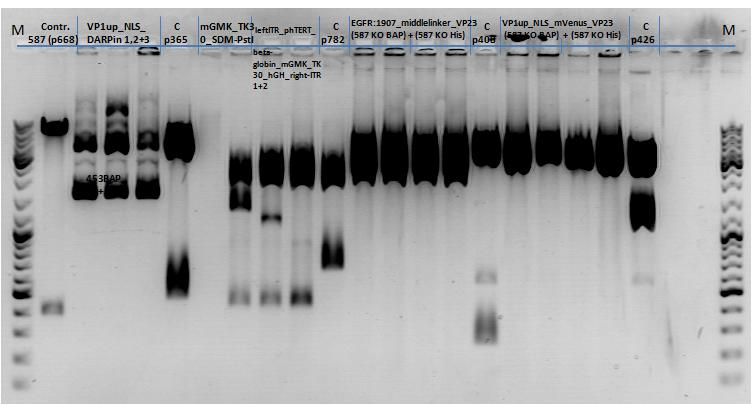
136. labday 01.10.2010
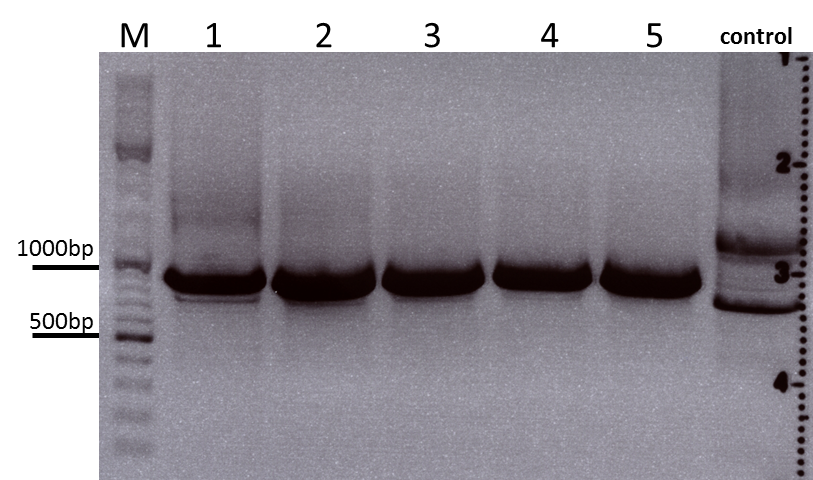
Test Digestions of yesterdays minipreps
Investigator: Achim, Anna
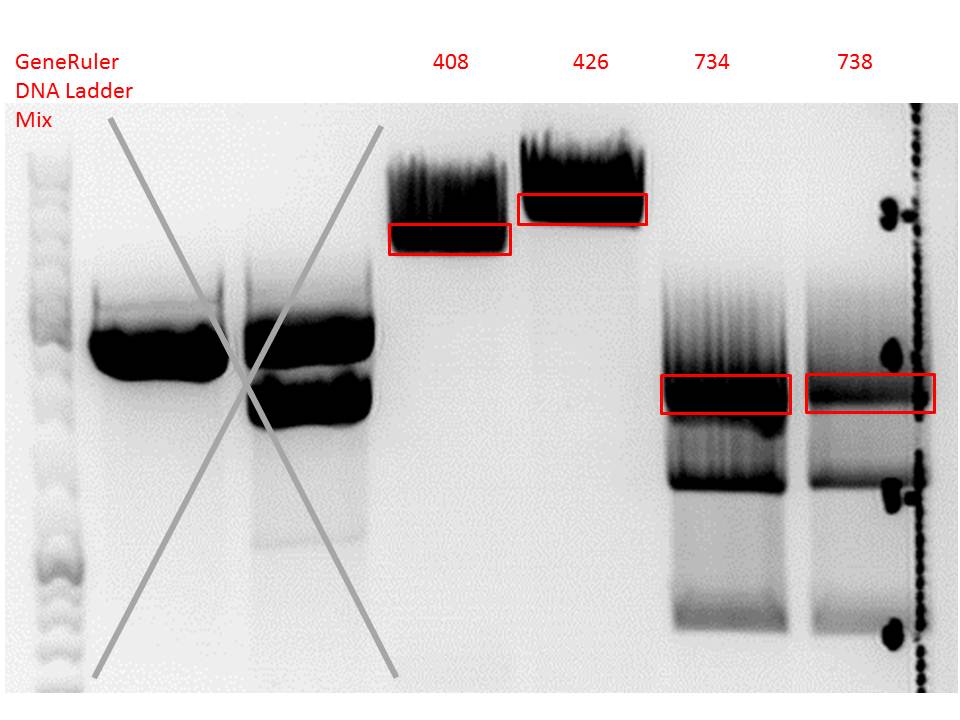
- 001_CMV: Cut with EcoRI, PstI & PvuII
- P654 (Lane 1)
- P655 (Lane 2)
- P686 (Lane 3)
- Control P320 (Lane 4)
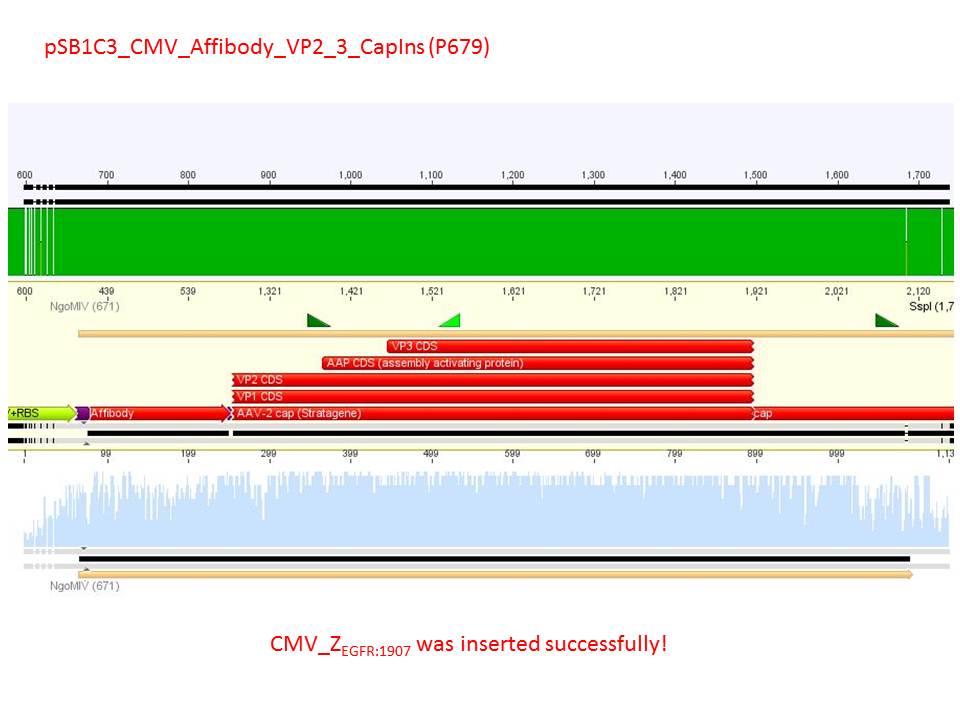
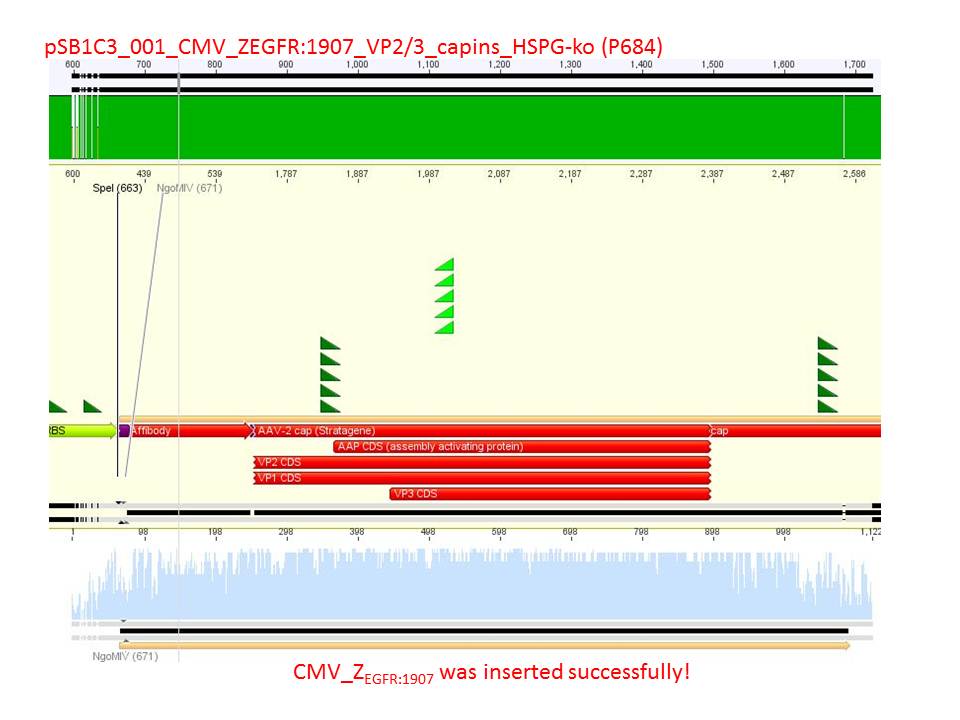
- CMV_ZEGFR:1907_VP2/3_inscap: Cut with EcoRI & SspI
- P678 (Lane 5)
- P679 (Lane 6)
- Control P514 (Lane 7)
- Results:The expected bands should run at 1700 and 3000 bp. P679 was sent for sequencing.
- CMV_ZEGFR:1907_VP2/3_inscap_HSPG-KO: Cut with EcoRI & SspI
- P680 (Lane 8)
- P681 (Lane 9)
- P684 (Lane 10)
- P685 (Lane 11)
- Control P613 (Lane 12)
- Results:The expected bands should run at 1700 and 3000 bp. P684 was sent for sequencing.
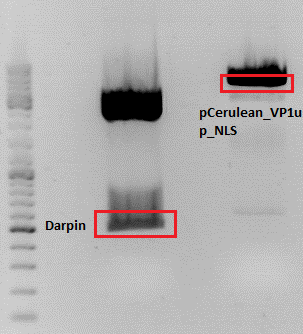
- pCerulean_Vp1up_NLS_Darpin: Cut with BamHI & XbaI
- P676 (Lane 13)
- P677 (Lane 14)
- P683 (Lane 15)
- Control P365 (Lane 20)
- Results:The expected bands should run at 500 and 4500 bp. However, just one band at ~3000 bp is visible on the gel.
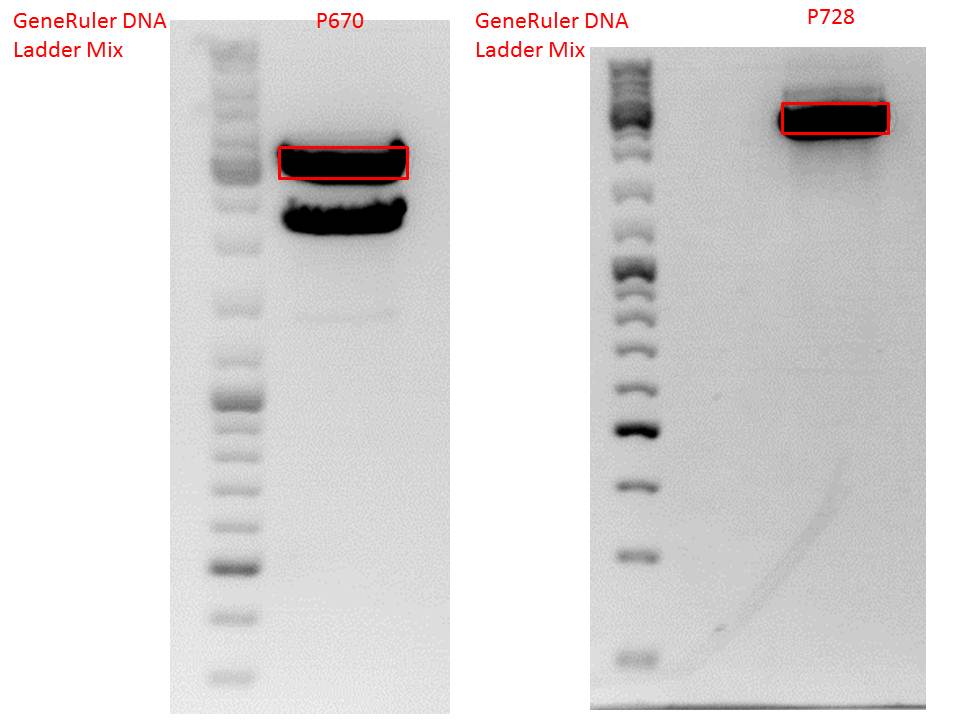
Samples Stefan: Cut with Xba & Spe
- P670 (Lane 16)
- P671 (Lane 17)
- P672 (Lane 18)
- P673 (Lane 19)

mini preps of CD clones
Investigator: Kira
c(p689)= 115 ng/ul
c(690)= 151, 20 ng/ul
c(691)= 122,27 ng/ul
c(p692) = 126,42 ng/ul
test digestion of CD clones
Investigator: Kira
| Components | sample Volume/µL |
| DNA | 4,0 µl |
| BSA (10x) | 2 µl |
| Buffer no. 4 | 2,0 µl |
| Enzyme 1 XbaI | 1,0 µl |
| Enzyme 2 AgeI | 1,5 µl |
| H2O | 9,5 µl |
| Total volume | 20 |
incubation @ 37 C for approx. 2 h
1% agarose gel
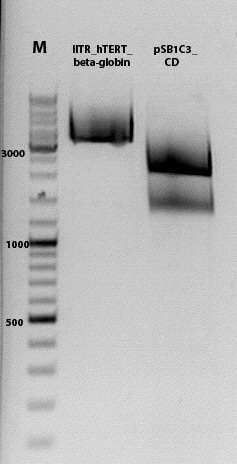
Biobrick assembly pSB1C3_lITR_hTERT_beta-globin_CD
Investigator: Kira
c(pSB1C3_lITR_hTERT_beta-globin)= 333 ng/ul
c(pSB1C3_CD)= 151 ng/ul
| Components | vector Volume/µL | insert Volume/µL |
| DNA | 4,5 µl | 6 |
| BSA (10x) | 3 µl | 3 |
| Buffer no. 4 | 3,0 µl | 3 |
| Enzyme 1 XbaI | 0 µl | 1,5 |
| Enzyme 2 SpeI | 1,5 µl | 0 |
| Enzyme 3 PstI-HF | 1,0 | 1 |
| H2O | 17 | 15,5 |
| Total volume | 25 |
incubation @ 37 C for approx. 2 h
1% agarose gel
Ligation
DNA-mix: 8 ul (vector 4,6ul)+(insert 3,4 ul)
T4 ligase: 1 ul
T4 buffer: 1 ul
Incubation @ RT for 30 min
Transformation was performed according to the standard protocol w BL21 cells.
Sequencing results of pCerulean_Zegfr:1907_MiddleLinker_VP2/3_HSPG-KO
Investigator: Hanna
Comment: All N-terminal fusion approaches with VP2/3_HSPG-KO revealed positive results except of pCerulean_Zegfr:1907_MiddleLinker_VP2/3_HSPG-KO. Another clone was picked, preped, test digested and sent for sequencing.
Conclusion: Sequencing results revealed positive results.
Picking clones of pGA14_MiddleLinker_VP2/3_insCap and pGA14_MiddleLinker_VP2/3_HSPG-KO
Investigator: Hanna
Unfortunately there grew a bacteria lawn over night - it was hardly not possible to pick clones. Nevertheless I tried and picked 2 clones of pGA14_MiddleLinker_VP2/3_insCap and pGA14_MiddleLinker_VP2/3_HSPG-KO.
To do: Mini-Prep and test digestion.
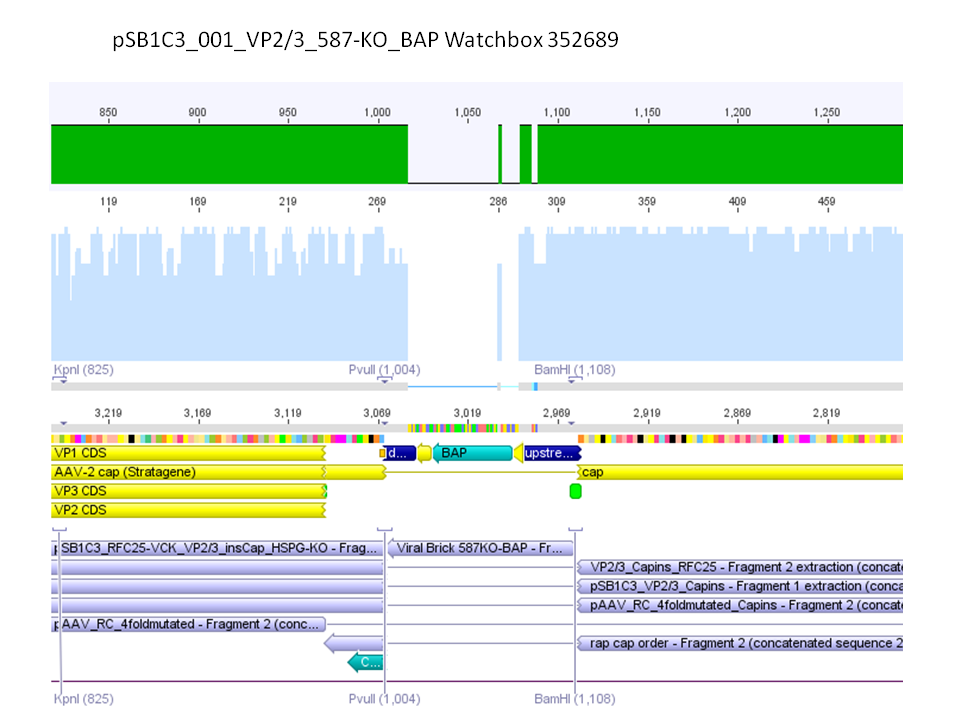
Sequencing results of pSB1C3_001_VP2/3_587-KO_BAP and pSB1C3_001_VP2/3_587-KO_6xHis
Investigator: Hanna
Comment: For the creation of our super constructs, the His-Tag and the BAP motif need to be cloned into VP2/3 for N-terminal fusion to VP2. Sequencing results showed that the 6xHis and BAP motif was not cloned into VP2/3_insCap.
To do: Clone 587-KO_6xHis and 587-KO_BAP into pSB1C3_001_VP2/3_insCap 1. via digestion of inserts and 2. via hybridization of referring oligos - digestion of vector with 800-900 ng.
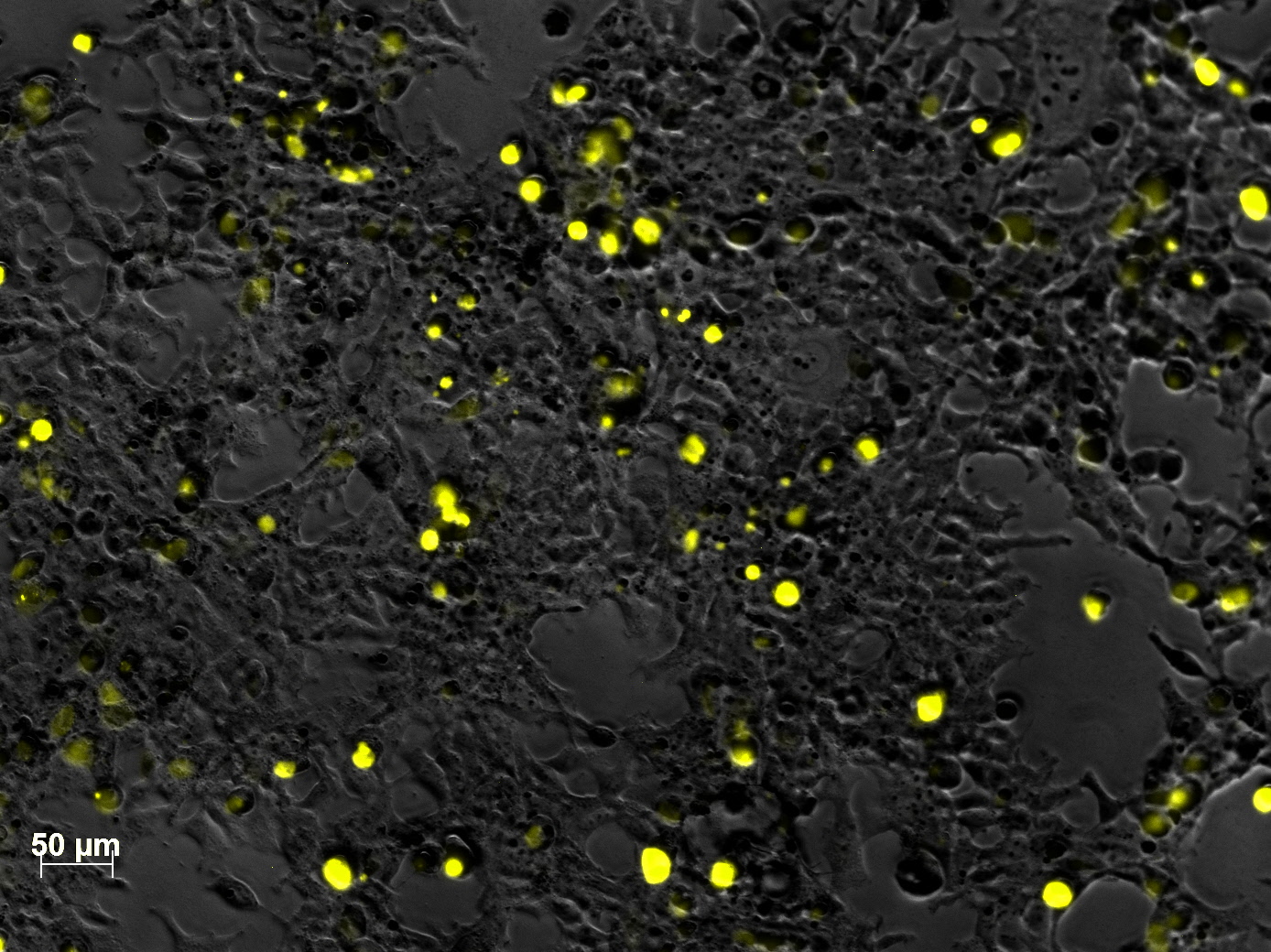
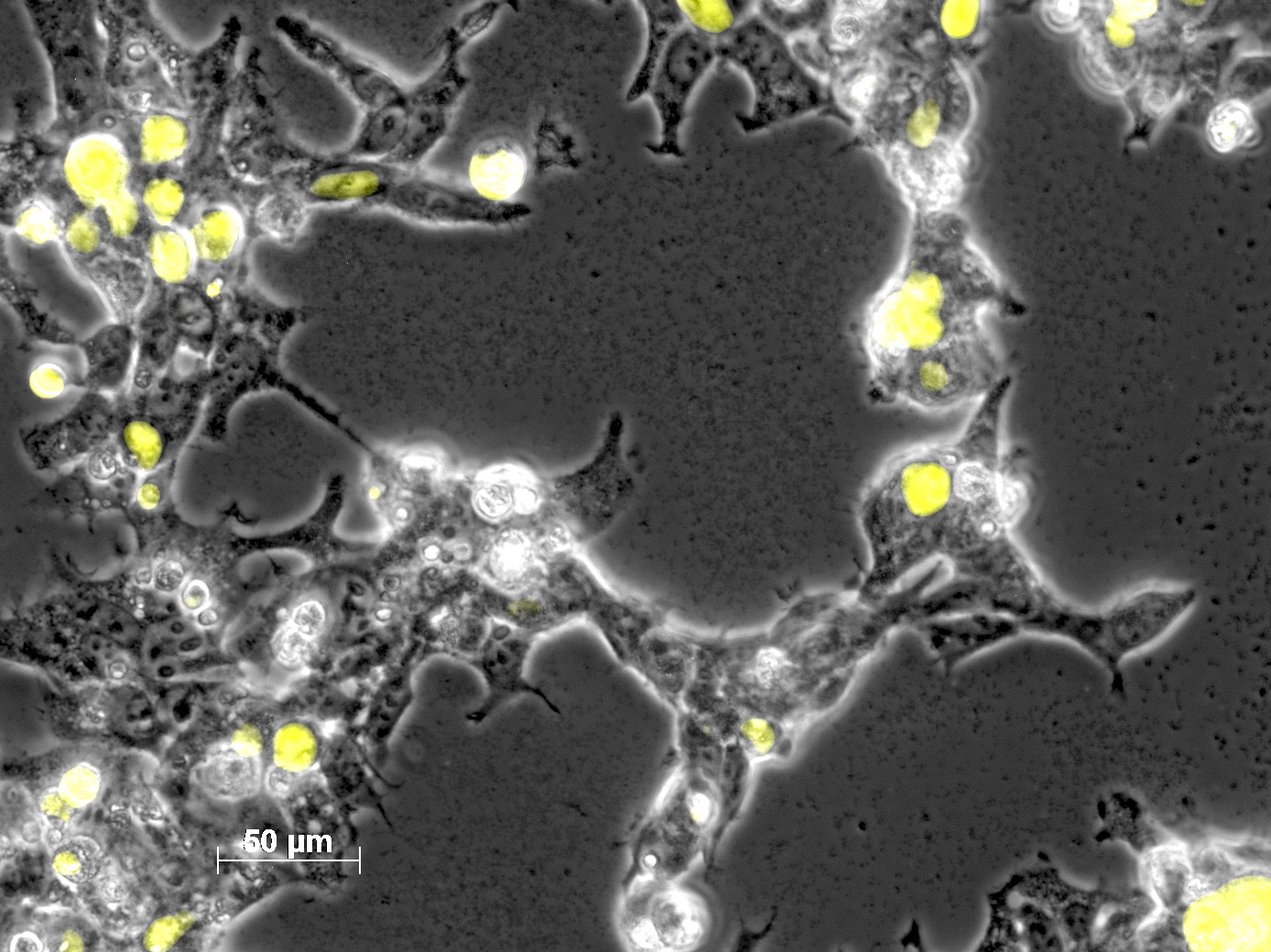
Impressions of transfection of AAV293 with pCerulean_VP1up_NLS_mVenus_VP2/3
Investigator: Adrian
Yesterday AAV293 cells were transfected with pCerulean_VP1up_NLS_mVenus_VP2/3 and GMK-TK30 as gene of interest. Today, nuclear localization of the produced mVenus_VP2/3 fusion protein was visible and can be nicely seen in the following pictures:
Conclusion: The VP2/3 fusion particle was transcribed and translated AND was transported back into the nucleus in order to be packaged by the ITR-flanked gene of interest.
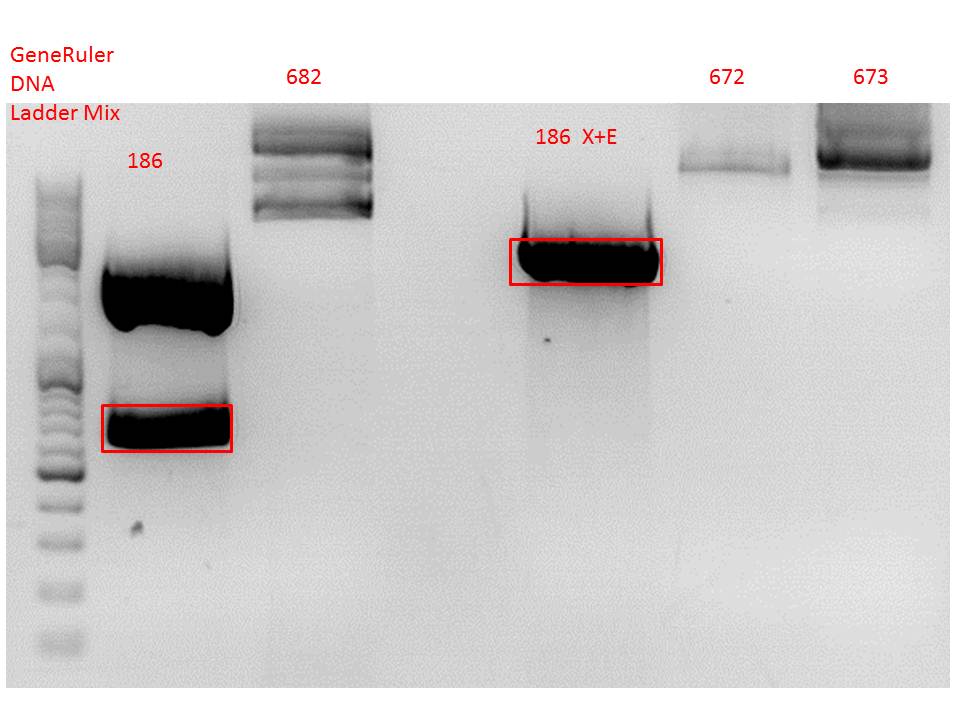
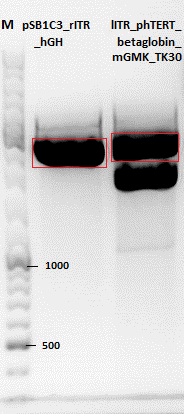
Cloning of hGH_rITR into pSB1C3_lITR_phTERT_beta-globin_CFP and lITR_phTERT_beta-globin_mGMK_TK30 into pSB1C3_hGH_rITR
Investigator: Stefan
Vector name:
- pSB1C3_lITR_phTERT_beta-globin_CFP (P682)
- pSB1C3_hGH_rITR (P186)
Insert name:
- pSB1C3_hGH_rITR (P186)
- pSB1C3_lITR_phTERT_beta-globin_mGMK_TK30 (P672)
- pSB1C3_lITR_phTERT_beta-globin_mGMK_TK30 (P673)
| components | volume for P186 /µl | volume of P186 X+E /µl | volume of P672 + P673 /µl | volume of P682 /µl |
| DNA | 12 | 12 | 5 | 14 |
| BSA (10x) | 2,5 | 2 | 2 | 2 |
| Buffer 4 (10x) | 3 | 2 | 2 | 2 |
| Enzyme XbaI | 1 | 1 | - | - |
| Enzyme PstI | 1 | - | - | 1 |
| Enzyme EcoI | - | 1 | 1 | - |
| Enzyme SpeI | - | - | 1 | 1 |
| H2O | 6 | 6 | 9 | - |
| Total volume (e.g. 15,20,25,30 µl) | 25 | 25 | 20 | 20 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
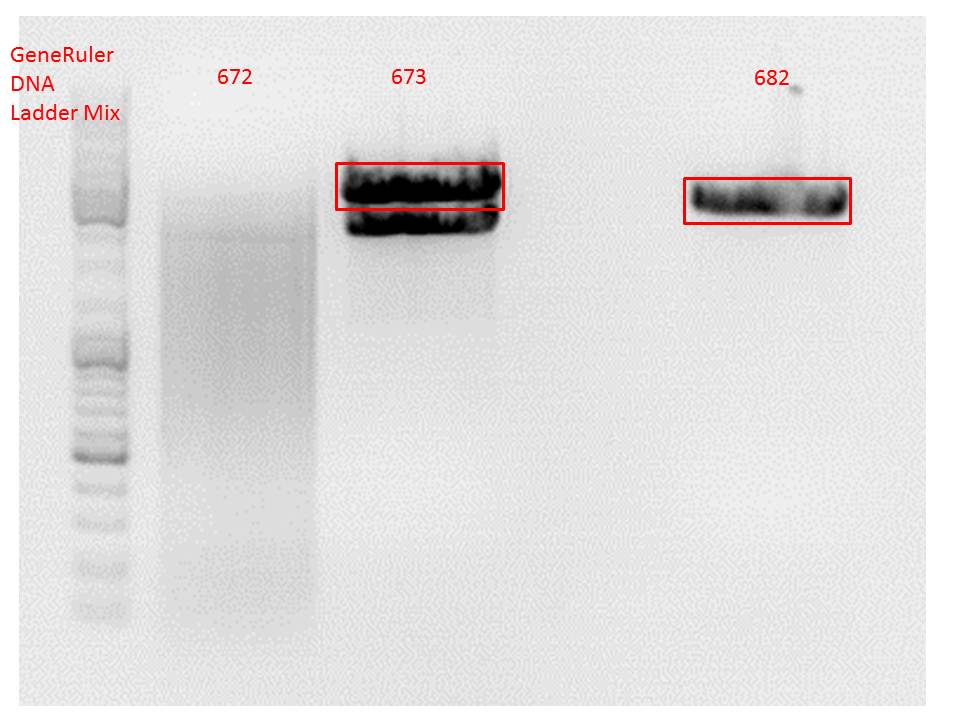
Comment: Since the first gel revealed problems with samples 672, 673 and 682 another digestion was prepared (same approach as shown above), this time using a 0,8% gel and another loading dye (without SDS).
0,4 g Agarose, 50 ml TAE (0,8%), 3 µl GELRED , at 115 Volt
Comment: Since sample 672 showed no bands this sample was discarded and cloning continued only using 673.
Gel extraction:
Was performed according to protocol.
T4 Ligation:
Ligation was performed over night.
| ligation name | 186 (X+E) + 673 | 186 + 682 |
| volume of vector | 3,51 | 4,49 |
| volume of insert | 5,38 | 2,64 |
Transformation:
Will be done tomorrow using BL21 cells.
New BL21 cells ready to use!
Investigator: Stefan
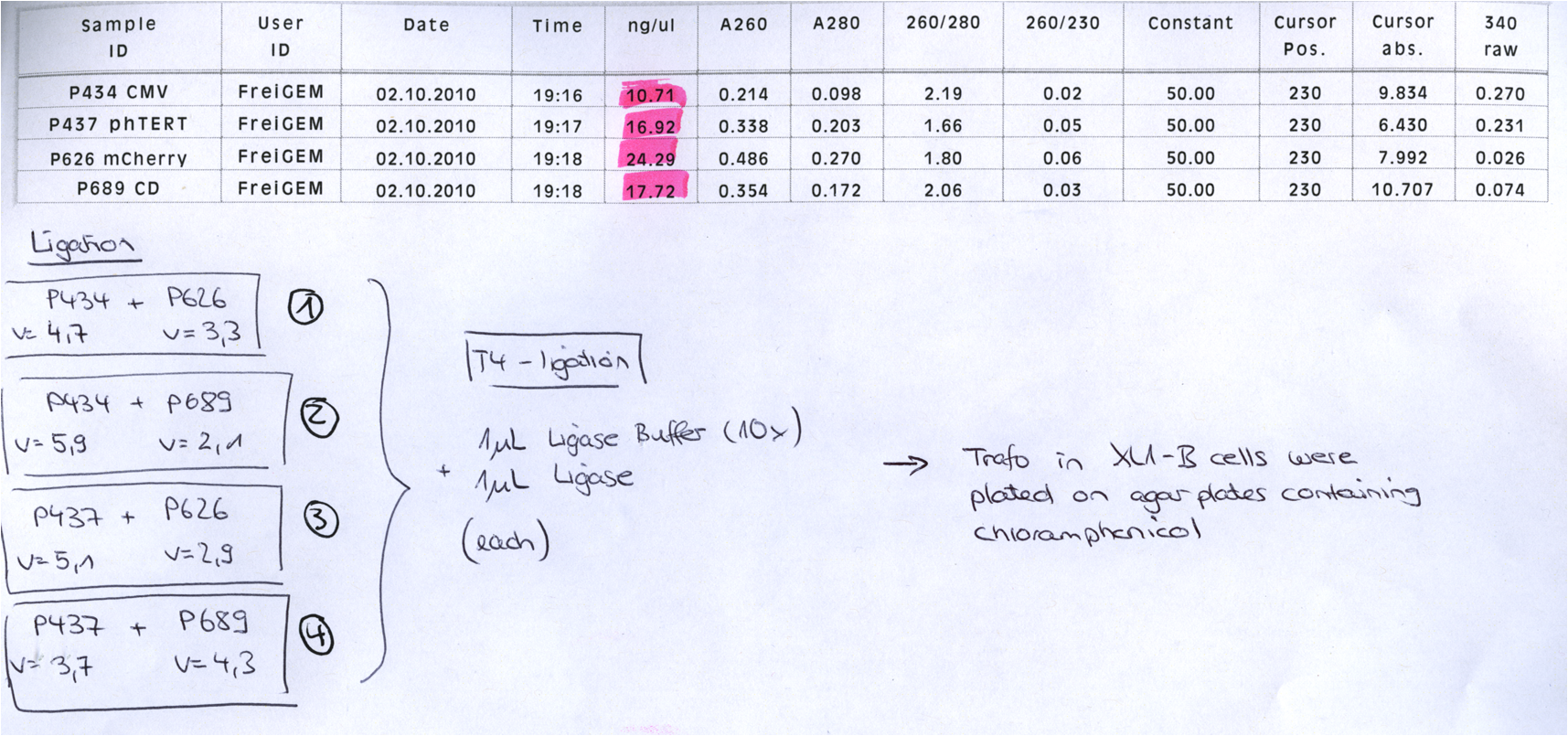
137. labday 02.10.2010
Repetition of the approach of subcloning the ViralBricks HSPG-ko BAP and His into VP2/3 with different strategies
Investigator: Bea
Comment: Unfortunately, the last experiment revealed that subcloning of the two ViralBricks did not work out which was shown by the sequencing results. Therefore we need to repeat the approach of subcloning the ViralBricks into the pSB1C3_VP2/3 which than can be used for fusing it to the n-terminal targeting approaches. Two different strategies will be carried out which means that we are digesting the pSB1C3_Viralbricks AND hybridize the oligos (BAP and His) in order to obtain some positive results.
The vector (P415) and the two ViralBricks BAP-ko and His-ko were digested with BamHi and PvuII. The vector was dephosphorylated following the standard protocol.
The vector was loaded on a 1% agarose gel before dephosphorylation was conducted. The results can be seen above in the gel picture:
Result:
The ligation was performed over night at 18°C with T4-ligase.
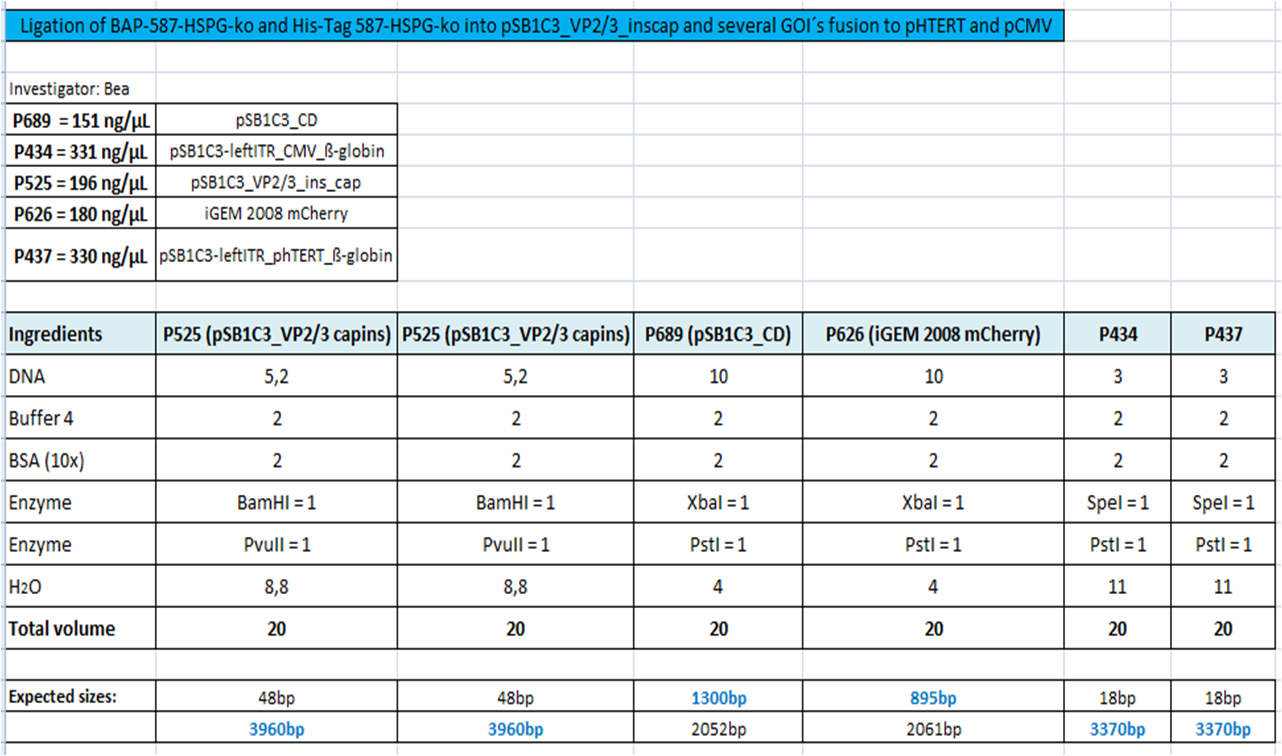
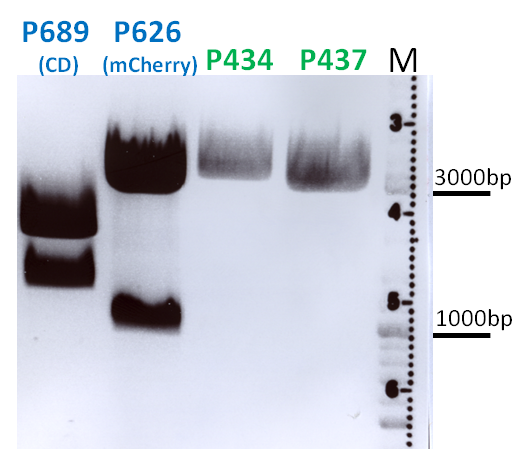
BioBrick Assembly of CD and mCherry
Investigator: Bea
Comment: We decided to choose some additional GOI´s for the vectorplasmid so the "new" GOI´s need to be fused to the leftITR_Promoter_betaglobin constructs. Since only one clone grew on the plate prepared by Kira where she fused the CD (cytosine deaminase) to the above mentioned construct I am going to repeat it aswell.
Digestion of the constructs:
- P689 = pSB1C3_CD
- P626 = iGEM_mCherry
- P434 = pSB1C3_leftITR_CMV_betaglobin
- P434 = pSB1C3_leftITR_phTERT_betaglobin
- P525 = pSB1C3_VP2/capins
Loading plan:
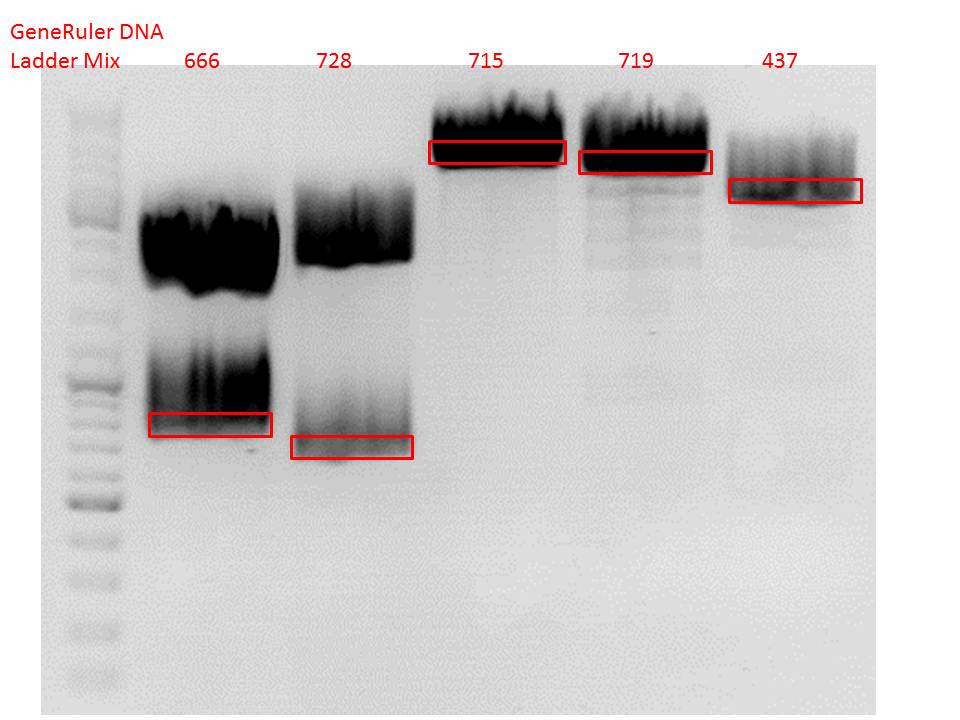
P689 (pSB1C3_CD) P626 (iGEM mCherry) P434 (pSB1C3_lITR_CMV_ß-globin) P437 (pSB1C3_lITR_phTERT_ß-globin)
Results:
After gel extraction has been performed, the ligation was carried out.
Ligation:
The ligation mix was transformed into XL1-Blue cells and plated on agar plates containing chloramphenicol.
Next steps:
Picking clones and perform Mini-Prep. After the Mini-Preps have been performed this construct needs to be sequenced and tested in cell culture.
comment: just one clone? uhhhh, when did you take out the plate? i put it at around 1 am..maybe they need a while to grow, no? (Kira)
Yep, I found another clone ;-) But the two look pretty good. I took them out today in the afternoon - I guess that is enough time! But if you want I can put them back into the 37° C room??! (Bea)
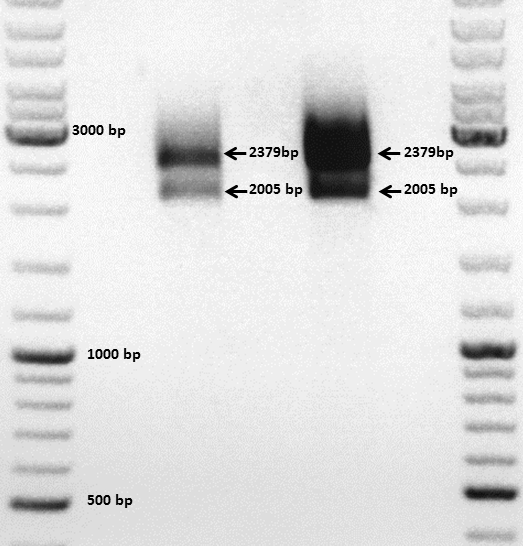
Mini Prep and test digestion of pGa14_MiddleLinker_VP2/3_HSPG-KO
Investigator: Hanna
Comment: In order to fuse the DARPin to the N-terminus of VP2/3_insCap and VP2/3_HSPG-KO, these motifs need to be fused to the Middle Linker (performed on 30.9.2010). In order to be able to immediately continue with cloning, BL21 cells were transformed and clones were picked in the noon. Now, the DNA needs to be preped and test digested in order to perform a 3-fragment-ligation with the DARPin and the pSB1C3_001_CMV plasmid. Unfortunately the cells of the VP2/3_insCap construct grew not densely enough. Therefore two new clones were picked from the plate and were inoculated in 10 mL LB medium. The other construct (pGa14_MiddleLinker_VP2/3_HSPG-KO) was preped and test digested.
TEST DIGESTION:
For both clones:
- DNA: 4 µL
- Buffer 4: 1 µL
- BSA: 1 µL
- EcoRI: 0.5 µL
- PstI: 0.5 µL
- H2O: 3 µL
Incubation: 1 hour.
Agarose gel: 0.8 %, 115 Volt, 1 hour.
Comment: The choice of these restriction enzymes was not clever, because if cloning didn't work, I expect one band at 2379 bp (the insert should hardly be seen) - if it worked one band at 2379 bp and a second at 2005 bp. Fortunately the bands could be separated ans revealed positive results for both clones :
Next steps:
- Mini-Prep and test digestion of pGa14_MiddleLinker_VP2/3_ins Cap.
- 3 fragment ligation of MiddleLinker_VP2/3_ins Cap or MiddleLinker_VP2/3_HSPG-KO + DARPin + pSB1C3_001_CMV backbone.
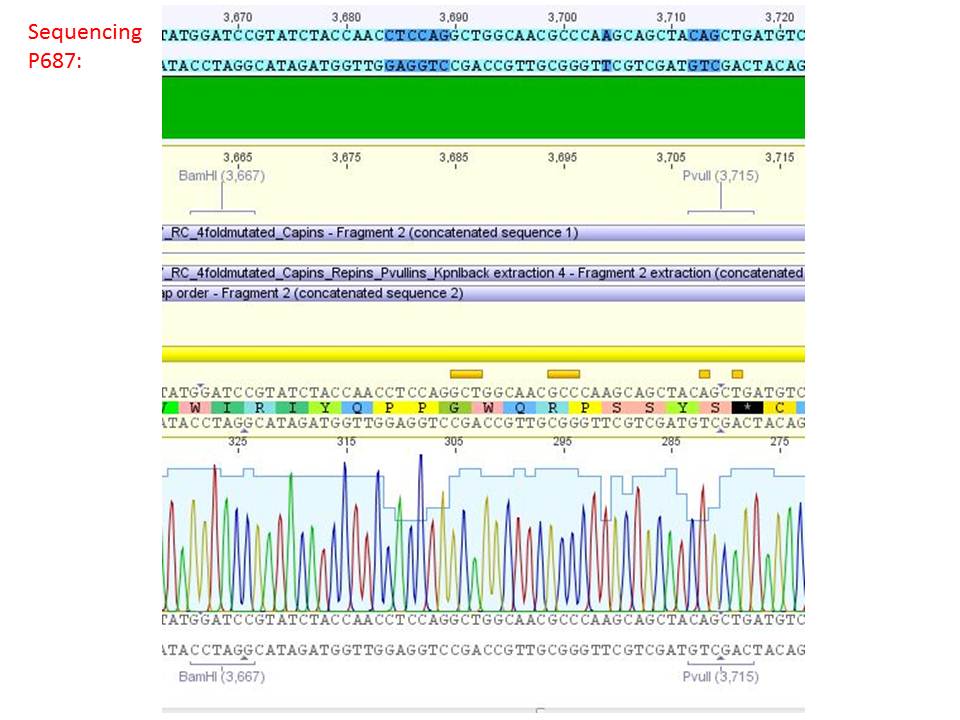
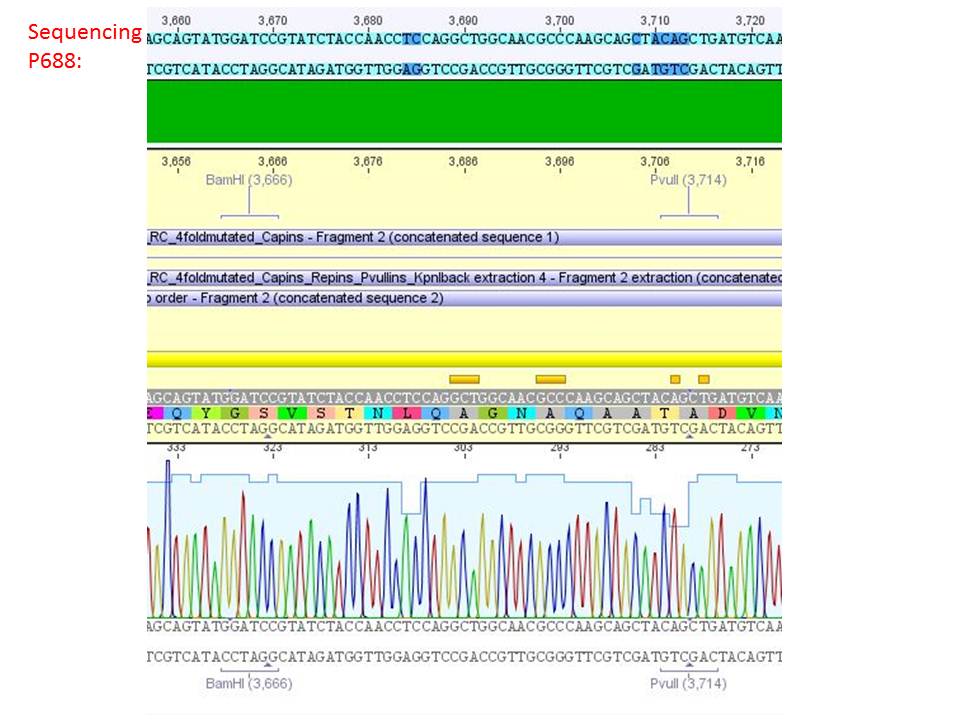
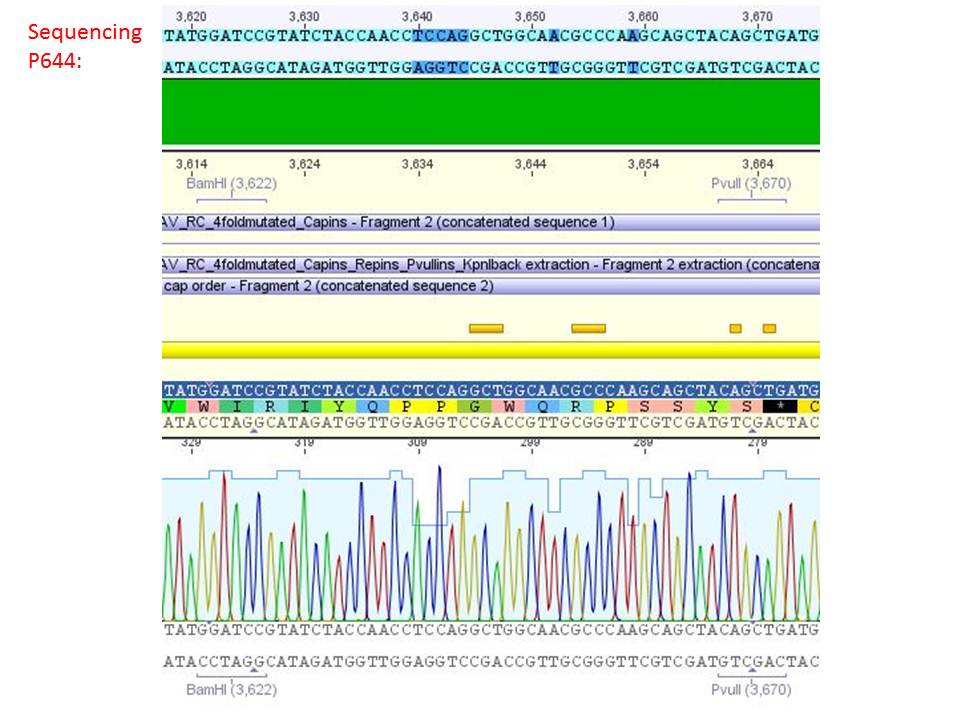
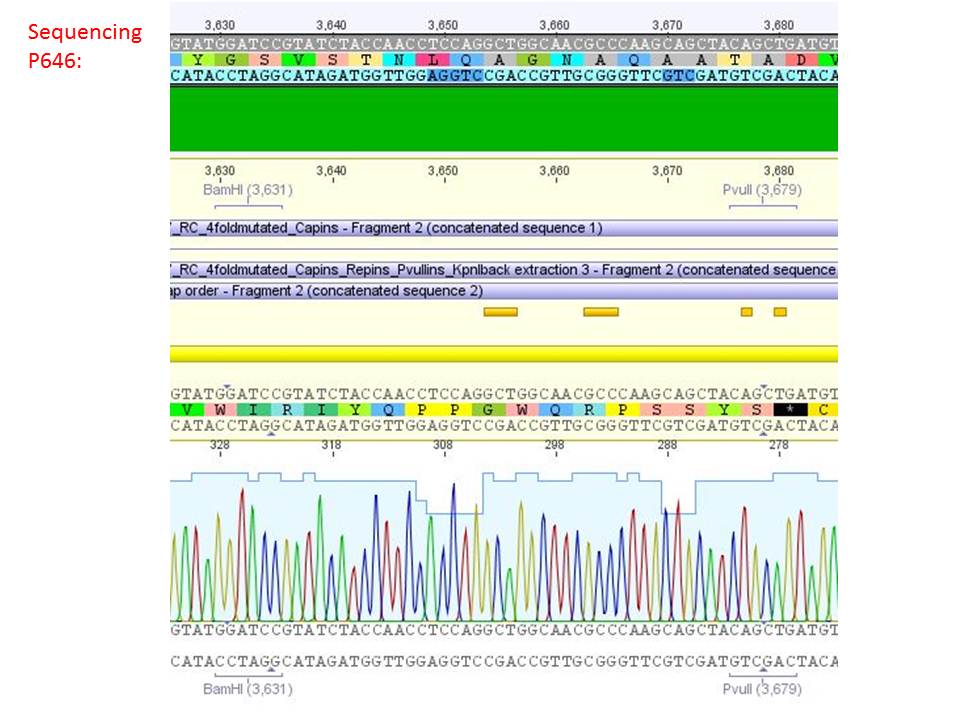
Sequencing analysis of HSPG-ko in several IRCK constructs
Investigator: Stefan
pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10 clone 1 (P687):
pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10 clone 2(P688):
pSB1C3_001_RC_IRCK_VP1-ko_HSPG-ko_P5tataless cl1 (P644):
pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1 (P646):
Comment: Sequencing results show that all plasmids contain the HSGP-ko. Working will be continued using P644, P646 and P687.
Mini Prep of pGa14_MiddleLinker_VP2/3_inscap and pSB1C3_Darpin
Investigator: Jessica
- pSB1C3_Darpin clone1 c=158.5 ng/µl
- pSB1C3_Darpin clone2 c=205.6 ng/µl
- pGA14_MiddleLinker_VP2/3_inscap clone 1 c=147.5 ng/µl
- pGA14_MiddleLinker_VP2/3_inscap clone 2 c=121.0 ng/µl
Test-digestion:
| Components | pSB1C3_Darpin clone1/2 |
| DNA | 1,5 µl |
| BSA (10x) | 1 µl |
| Buffer no. 4 | 1 µl |
| Enzyme 1 XbaI | 0,4 µl |
| Enzyme 2 AgeI | 0,4 µl |
| H2O | 5,7 µl |
| Total volume | 10 |
Hanna wrote that a 3rd enzyme is need for the test digestion of pGA14_MiddleLinker_VP2/3_inscap but I couln't find any sequence of pGA14_MiddleLinker_VP2/3_inscap or pGA14

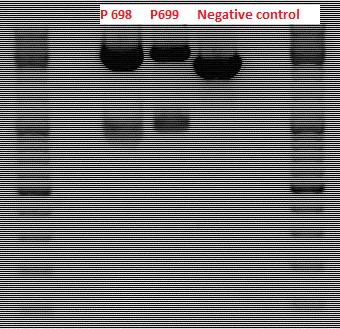
Test digestion of pGA14_MiddleLinker_VP2/3_inscap
Investigator: Achim
- pGA14_MiddleLinker_VP2/3_inscap clone 1 c=147.5 ng/µl (P698)
- pGA14_MiddleLinker_VP2/3_inscap clone 2 c=121.0 ng/µl (P699)
- Control: pGA14_MiddleLinker (P301)
- Digestion with EcoRI & SalI; should yield two bands of 3400 & 1000 bp. The negative control should only be cut once.
Mass Midi-Prep III
Investigator: Chris W.
Midi-Prep of:
pSB1C3_001_RC_IRCK_VP1-ko_HSPG-ko_P5tataless cl1 =P700 =B521
pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1 =P701 =B523
pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10 clone 1 =P702 =B561
pCerulean_ZEGFR:1907_MiddleLinker_VP2/3_HSPG-KO clone 2 =P703 =B543
pCerulean_CFP_MiddleLinker_VP2/3_HSPG-KO =P704 =B507
pCerulean_ZEGFR:1907_SEG_VP2/3_HSPG-KO =P705 =B509
pCerulean_ZEGFR:1907_ShortLinker_VP2/3_HSPG-KO =P706 =B510
pCerulean_ZEGFR:1907_LongLinker_VP2/3_HSPG-KO =P707 =B511
pCerulean_6xHis_MiddleLinker_VP2/3_HSPG-KO =P708 =B512
pCerulean_VP1up_NLS_ZEGFR:1907_VP2/3_HSPG-KO =P709 =B513
pCerulean_VP1up_NLS_6xHis_VP2/3_HSPG-KO =P710 =B514
pCerulean_VP1up_NLS_mVenus_VP2/3_HSPG-KO =P711 =B515
pSB1C3_001_RC_IRCK_P5tataless clone 1 =P712 =B516
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P700 | P701 | P702 | P703 | P704 | P705 | P706 | P707 | P708 | P709 | P710 | P711 | P712 |
| concentration (ng/µl) | 695 | 370 | 601 | 1548 | 3996 | 3630 | 2460 | 2610 | 1860 | 2913 | 2964 | 3030 | 1200 |
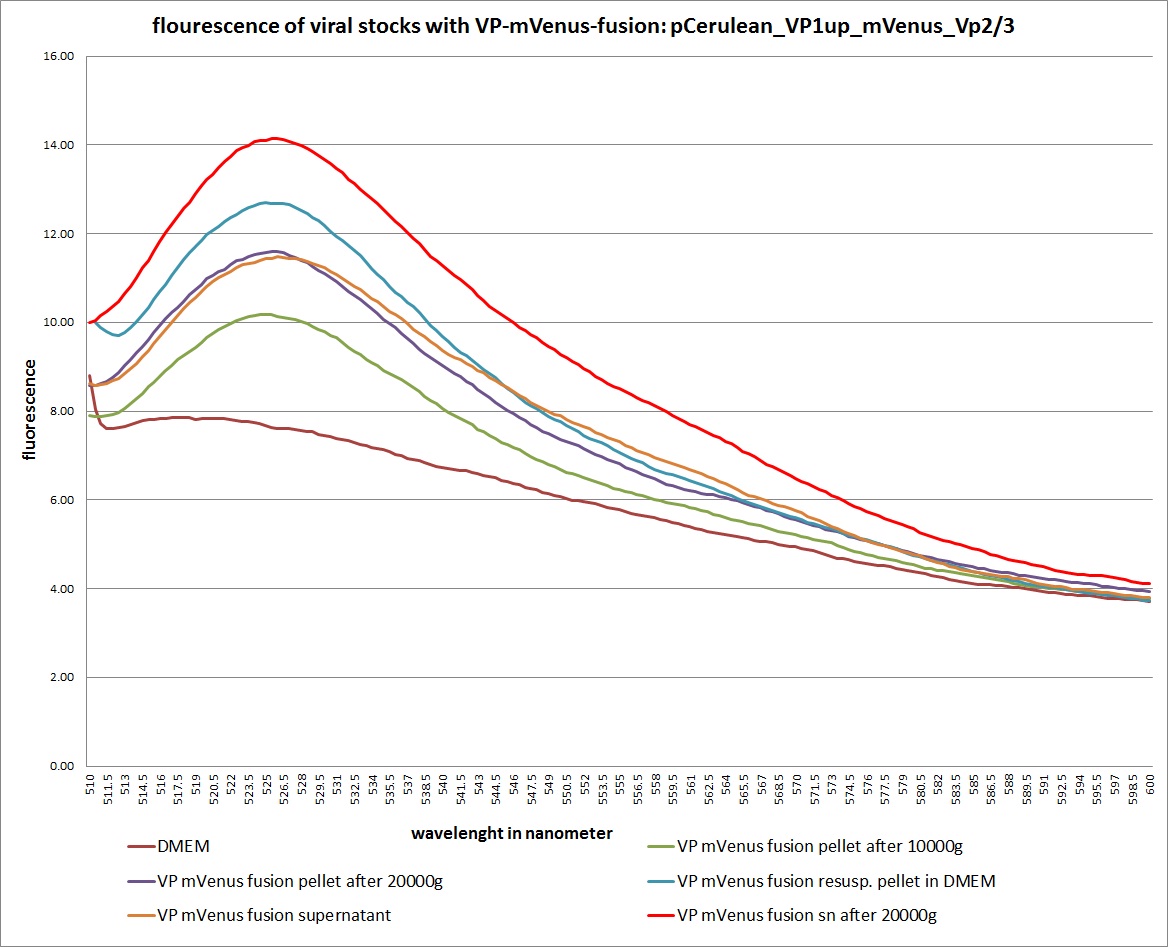
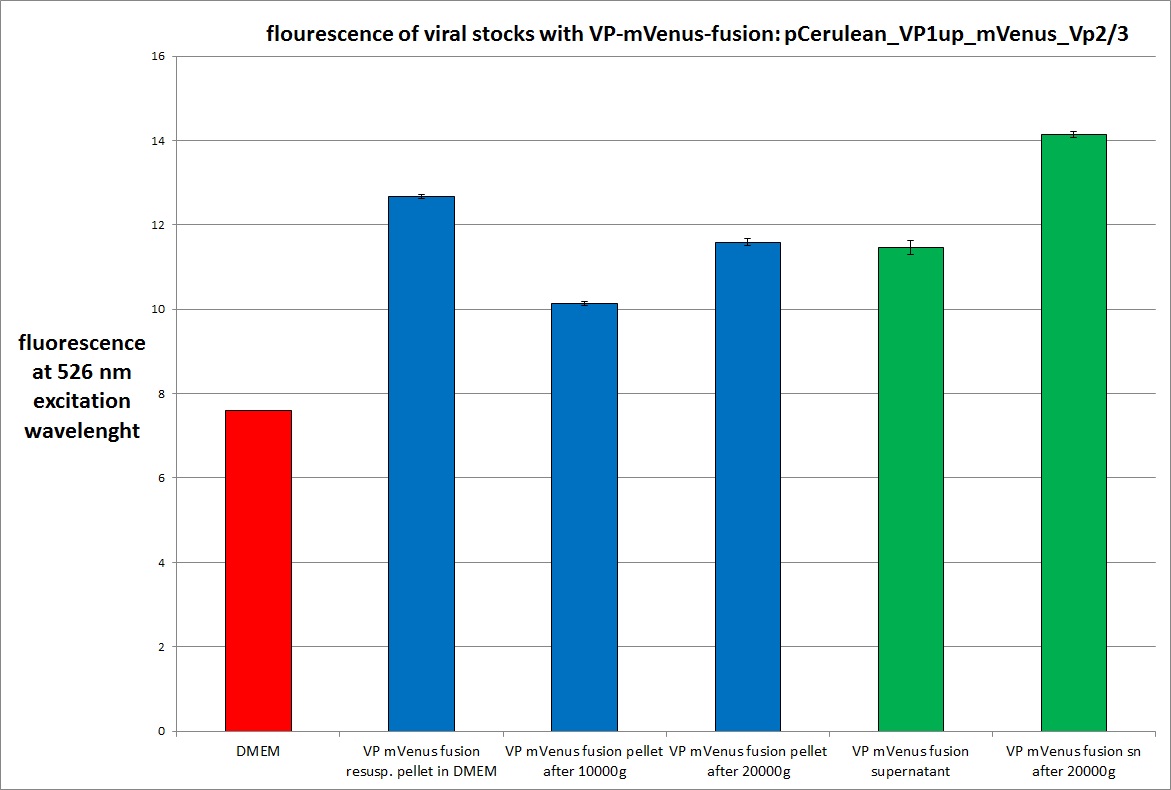
Analysis of viral harvest via spectrometer
Investigator: Adrian, Patrick
The motivation: We want to figure out if it is possible to purify our viral stocks via centrifugation with 10.000 g and/or 20.000 g. In theory/ according to the literature the viral particles should be in the cells and attached to the HSPGs at the cellular surfaces.
The plan: Two different viral stocks were prepared:
Stock 1 with VP2-mVenus-fusion-Capsid loaded with TKGMK (4 wells from a 6 well plate)
- R/C 1: 50% pCerulean_VP1up_NLS_mVenus_Vp2/3 P501
- R/C 2: 50% Rep/Cap2: P449 pAAV_RC_4fachmut_VP1-ko:
- GOI: TKGMK: P82.b
- pHelper
Stock 2 "standard" virus (4 wells from a 6 well plate)
- R/C 1 P486
- GOI: TKGMK: P82.b
- pHelper
These two stocks got harvested according to standard protocol (scratching cells, transfer them in to 15 ml falcons, resuling 12ml solution total). After centrifugation (10 min 200g), the supernatant got transferred into two new 15 ml falcons and the pellets were resuspended with 10 ml DMEM Medium. The Stocks got freezed thawed two times so finally there were:
- DMEM as negative control
- resuspended Pellet with mVenus-fusioned viral capsids
- resuspended Pellet with "standard virus"
- supernatant with mVenus-fusioned viral capsids
- supernatant with "standard virus"
The fluorescence of each stock was measured via spectrometer. After that, stocks 2 and 4 were centrifugated 10min with 10.000 g followed by an other fluorescence measuring. Finally the stocks got spin down with 20.000 g for 10 min
The results:
The conclusions:
The highest amount of fluorescence was measured in the supernatant, the individual centrifugation steps had no decrease in fluorescence as consequence!
Keep in mind, without further investigation it is not valid to say that we can purify our stocks via 10-20.000 g centrifugation steps, because we dont know if the viral capsids are still intact! The FACS analysis are not sufficient enough to make a decision, because our last samples had YFP as GOI (=> so efficient cell sorting was not possible). We need to do qPCRs to make a valid evidence.
200µl-Transduction for testing the pTERT-promoter
Investigator: Adrian, Patrick
We transduced A431, HT1080 and AAV293 cells with 4 different viral stocks:
- control vector from 15.9, 750.000 cells, 3.3 µg approach
- P25 R/C= 326 pTERT_mVenus
- P26 pAAV_RC_RepCapIns_SDMKpnI clone 1 P431 pTERT_mVenus
- His-linker-fusion
CRUCIAL: There was not enough viral stock: so the whole Transduction was performed with 200µl instead of 1000µl
There are 45 samples to FACS on monday 4.10. Tripple values for each stock of each cell lines.
Transfection
Investigators: Adrian, Patrick
Transfection followed standard protocol (3.3 µg of each plasmid, 1 ml 0.3M CaCl2 and 1 ml of 2xHBS buffer PH 11.12) but with 20 min incubation time (Ca-DNA-2xHBS)
following stocks:
- mVenus standard stock 10 x cm R/C: P4682 plates
- TKGMK stock 10 x cm2 plates R/C: P468
- viral particles with GOI (mVenus) missing HGH one 6well plate
- viral particles with GOI (mVenus) missing beta globin one 6well plate
- viral particles with R/C missing HSPG binding motif (P687) one 6well plate
- viral particles with new standard R/C (P668) one 6well plate
138. labday 03.10.2010
Proceeding with the approach of subcloning the ViralBricks HSPG-ko BAP and His into VP2/3 with different strategies
Investigator: Bea
Comment: Ligation was carried out over night and transformation will be performed today.
Since the digested vector was not sufficient for all four ligation I decided to perform the ligation only with the hybridized oligos 587-ko His and the digested Viralbricks 587-HSPG-ko His and BAP. If the digested BAP insert reverals no positive results, the hybrizied oligos for BAP can be used for another approach. The transformation was carried out in
- BL-21 cells
- and plated on agar plastes containing chloramphenicol
Midi-Prep of pSB1C3_lITR_CMV_betaglobin_mVenus_hGH_rITR clone1
Investigator: Chris W.
Midi-Preps of pSB1C3_lITR_CMV_betaglobin_mVenus_hGH_rITR clone1 =P713 =B200
The Midi-Prep were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P713 |
| concentration (ng/µl) | 1248,3 |
139. labday 04.10.2010
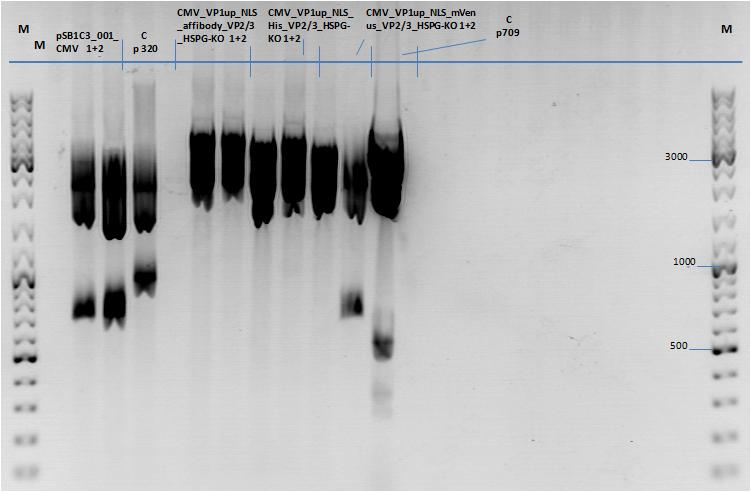
Three fragment ligation: pSB1C3_001_CMV_VP1up_NLS_Targeting_HSPG-KO_VP2/3
Investigator: Achim
Four ligations in total:
- Cloning CMV & VP1up_NLS_mVenus_HSPG-KO_VP2/3 into pSB1C3_001
- Cloning CMV & VP1up_NLS_His_HSPG-KO_VP2/3 into pSB1C3_001
- Cloning CMV & VP1up_NLS_affibody_HSPG-KO_VP2/3 into pSB1C3_001
- Cloning CMV into pSB1C3_001
Used plasmids:
- pSB1C3_CMV (P145)
- c=225.5 ng/µl -> 8 µl
- Cut with EcoRI & SpeI
- pCerulean_VP1up_NLS_mVenus_HSPG-KO_VP2/3 (P711)
- c=3030 ng/µl -> 0.7 µl
- Cut with XbaI & PstI
- pCerulean_VP1up_NLS_His_HSPG-KO_VP2/3 (P710)
- c=2965 ng/µl -> 0.7 µl
- Cut with XbaI & PstI
- pCerulean_VP1up_NLS_affibody_HSPG-KO_VP2/3 (P709)
- c=2913 ng/µl -> 0.7 µl
- Cut with XbaI & PstI
- pSB1C3_001_BLA (P320)
- c=408.5 ng/µl -> 5 µl
- Cut with EcoRI & PstI for three fragment ligations
- Cut with EcoRI & SpeI for ligation with CMV
´Seeding AAV293 for Transfection
Investigator: Patrick
9 6-well plates were prepared for the transfection tomorrow. 480000 cells were seeded into each well.
FACS Analysis of pTERT_mVenus-loaded-viral particles and pCerulean_VP1up_His_VP2/3
Investigators: Kerstin, Anissa
45 samples of the three cell lines: AAV293, A431 and HT1080 got transduced (see 2.10) with following viral stocks:
1.
- pHelper
- R/C : 326
- pTERT-mVenus
2.
- pHelper
- R/C : P431
- pTERT-mVenus
3.
- pHelper
- R/C P158a
- CMV_mVenus
4.
- pHelper
- R/C: pCerulean_VP1up_His_VP2/3
- pAAV_RC_4fachmut_VP1-ko
- CMV_mVenus
Assembly of pSB1C3_CMV_DARPin_VP2/3_insCap and pSB1C3_CMV_DARPin_VP2/3_HSPG-KO
Investigators: Hanna
Comment: We also want to test the DARPin_E01 for the N-terminal fusion approaches. Today a 3 fragment ligation will be performed in order to complete the VP2 fusion constructs with the DARPin.
Because 2 test digestions showed that the fusion of the DARPin to VP1up_NLS didn't work as ecpected (insert is too large) this has to be repeated before also the VP1 insertion with the DARPin can be completed.
Digestion
| components | volume of pSB1C3_CMV /µl | volume of pG14_ML_VP2/3_insCap /µl | volume of pG14_ML_VP2/3_HSPG-KO /µl | volume of pSB1C3_001_DARPin /µl |
| DNA | 9 | 6.8 | 10 | 5 |
| BSA (10x) | 2 | 3 | 3 | 2 |
| Buffer 4 (10x) | 2 | 3 | 3 | 2 |
| SpeI | 1 | - | - | - |
| PstI | 1 | 1 | 1 | - |
| NgoMIV | - | 1 | 1 | - |
| XmnI | - | 1 | 1 | - |
| XbaI | - | - | - | 4 |
| H2O | 5 | 14.2 | 11 | 9 |
| Total volume | 20 | 30 | 30 | 20 |
- Incubation: 2.5 h
Agarose-Gel:
0.5 g Agarose, 50 mL TAE (1 %), 3 µL GELRED, at 115 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 |
|---|---|---|---|
| CMV | 20 µl | 4 µl | ~ 2700 bp |
| MiddleLinker_VP2/3_insCap | 30 µl | 4 µl | ~ 2000 bp |
| MiddleLinker_VP2/3_HSPG-KO | 30 µl | 4 µl | ~ 2000 bp |
| DARPin | 20 µl | 4 µl | ~ 475 bp |
Gelex
- CMV: 48.99 ng/µL
- MiddleLinker_VP2/3_insCap: 12.95 ng/µL
- MiddleLinker_VP2/3_HSPG-KO ng/µL
- DARPin: 9.59 ng/µL
Ligation
| Construct | CMV (µl) | MiddleLinker_VP2/3_insCap (µl) | DARPin (µl) |
| pSB1C3_CMV_DARPin_MiddleLinker_VP2/3_insCap | 1 | 3.5 | 3.5 |
| Construct | CMV (µl) | MiddleLinker_VP2/3_HSPG-KO (µl) | DARPin (µl) |
| pSB1C3_CMV_DARPin_MiddleLinker_VP2/3_HSPG-KO | 1 | 3.5 | 3.5 |
Trafo
Was performed following the standard protocol.
Cells: XL1b
Miniprep of serveral constructs
Investigator: Stefan
Glycerol stocks were prepared:
B573 pSB1C3_lITR_CMV_betaglobin_mCherry clone1
B574 pSB1C3_lITR_CMV_betaglobin_mCherry clone2
B575 pSB1C3_lITR_CMV_betaglobin_CD clone1
B576 pSB1C3_lITR_CMV_betaglobin_CD clone2
B577 pSB1C3_lITR_phTERT_betaglobin_mCherry clone1
B578 pSB1C3_lITR_phTERT_betaglobin_mCherry clone2
B579 pSB1C3_lITR_phTERT_betaglobin_CFP_hgH_rITR clone 1
B580 pSB1C3_lITR_phTERT_betaglobin_CFP_hgH_rITR clone 2
B581 pSB1C3_lITR_phTERT_betaglobin_CFP_hgH_rITR clone 1
B582 pSB1C3_lITR_phTERT_betaglobin_CFP_hgH_rITR clone 2
B583 pSB1C3_lITR_phTERT_betaglobin_CD clone 1
B584 pSB1C3_lITR_phTERT_betaglobin_CD clone 2
B585 pSB1C3_lITR_phTERT_betaglobin_CD clone 3
B586 pSB1C3_001_VP2/3_capins_587_KO_His_clone1
B587 pSB1C3_001_VP2/3_capins_587_KO_His_clone2
B588 pSB1C3_001_VP2/3_capins_587_KO_His_clone3
B589 pSB1C3_001_VP2/3_capins_587_KO_His_oligo_clone1
B590 pSB1C3_001_VP2/3_capins_587_KO_His_oligo_clone2
B591 pSB1C3_001_VP2/3_capins_587_KO_His_oligo_clone3
B592 pSB1C3_001_VP2/3_capins_587_KO_Bap_clone1
B593 pSB1C3_001_VP2/3_capins_587_KO_Bap_clone2
B594 pSB1C3_001_VP2/3_capins_587_KO_Bap_clone3
Mini-Prep was performed according to the standard protocol
P714 pSB1C3_lITR_CMV_betaglobin_mCherry clone1 c = 89,3 ng/µl
P715 pSB1C3_lITR_CMV_betaglobin_mCherry clone2 c = 312,4 ng/µl
P716 pSB1C3_lITR_CMV_betaglobin_CD clone1 c = 254,4 ng/µl
P717 pSB1C3_lITR_CMV_betaglobin_CD clone2 c = 314,8 ng/µl
P718 pSB1C3_lITR_phTERT_betaglobin_mCherry clone1 c = 409,8 ng/µl
P719 pSB1C3_lITR_phTERT_betaglobin_mCherry clone2 c = 322,1 ng/µl
P720 pSB1C3_lITR_phTERT_betaglobin_CFP_hgH_rITR clone 1 c = 265,1 ng/µl
P721 pSB1C3_lITR_phTERT_betaglobin_CFP_hgH_rITR clone 2 c = 238,8 ng/µl
P722 pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hgH_rITR clone 1 c = 145,9 ng/µl
P723 pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hgH_rITR clone 2 c = 100,5 ng/µl
P724 pSB1C3_lITR_phTERT_betaglobin_CD clone 1 c = 176,9 ng/µl
P725 pSB1C3_lITR_phTERT_betaglobin_CD clone 2 c = 165,0 ng/µl
P726 pSB1C3_lITR_phTERT_betaglobin_CD clone 3 c = 149,0 ng/µl
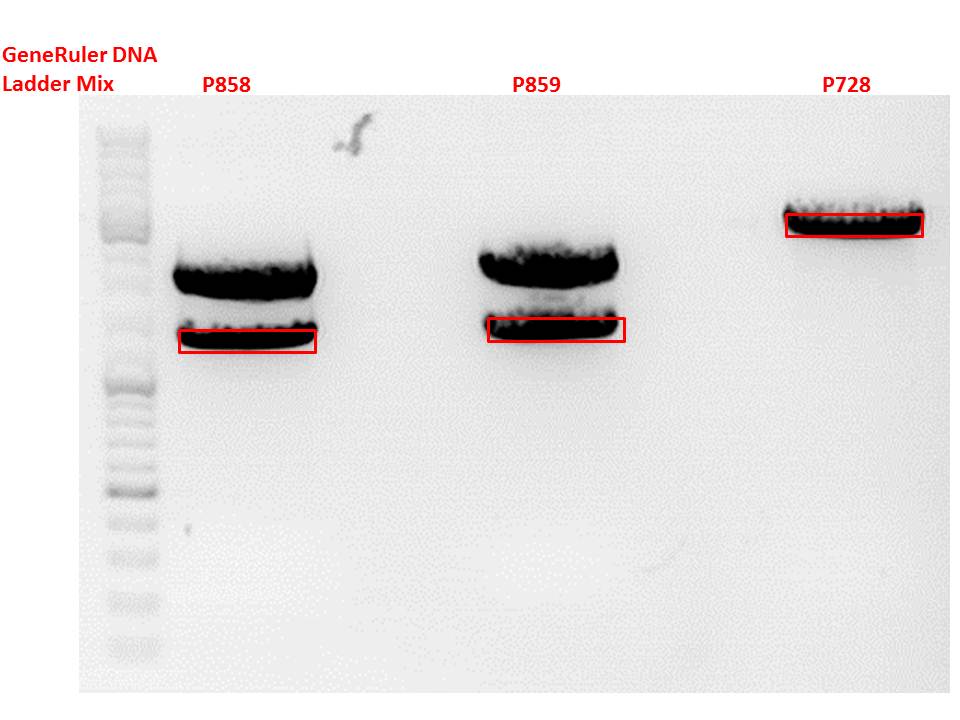
P727 pSB1C3_CMV c = 225,5 ng/µl
P728 pSB1C3_hGH_rITR c = 129,6 ng/µl
P729 pSB1C3_leftITR_CMV_beta-globin c = 243,4 ng/µl
P730 pSB1C3_leftITR_hTERT_beta-globin c = 81,8 ng/µl
P731 pSB1C3_001_VP2/3_insCap c = 466,1 ng/µl
P732 pSB1C3_001_VP2/3_capins_587_KO_His_clone1 c = 107,6 ng/µl
P733 pSB1C3_001_VP2/3_capins_587_KO_His_clone2 c = 110,3 ng/µl
P734 pSB1C3_001_VP2/3_capins_587_KO_His_clone3 c = 121,7 ng/µl
P735 pSB1C3_001_VP2/3_capins_587_KO_His_oligo_clone1 c = 101,0 ng/µl
P736 pSB1C3_001_VP2/3_capins_587_KO_His_oligo_clone2 c = 101,6 ng/µl
P737 pSB1C3_001_VP2/3_capins_587_KO_His_oligo_clone3 c = 87,0 ng/µl
P738 pSB1C3_001_VP2/3_capins_587_KO_Bap_clone1 c = 106,5 ng/µl
P739 pSB1C3_001_VP2/3_capins_587_KO_Bap_clone2 c = 110,2 ng/µl
P740 pSB1C3_001_VP2/3_capins_587_KO_Bap_clone3 c = 71,46 ng/µl
P741 pGGTBT7-ZEGFR:1907_Middlelinker_EGFP_His c = 115,4 ng/µl
140. labday 05.10.2010
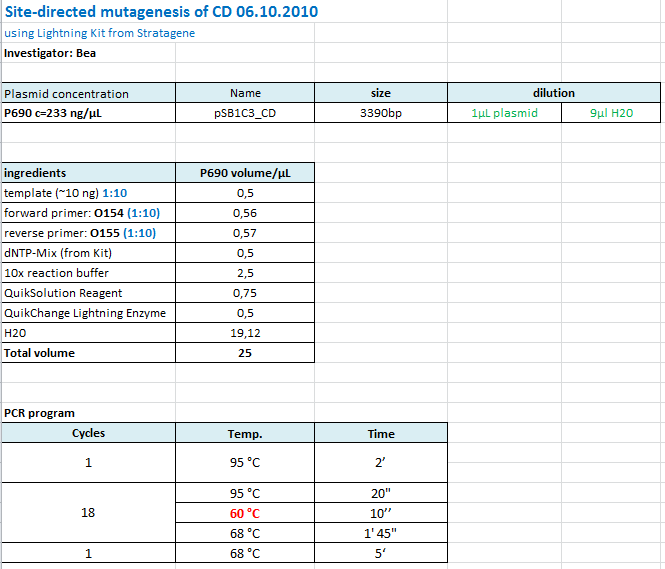
Site-directed mutagenesis in pSB1C3_mGMK_TK30 construct
Investigator: Bea
Comment: Since we are still waiting for the ordered TK30, idea was to mutate the PstI site in the TK30 region. After mutating the PstI it is ready for submitting. Addtionally the ordered and received sr3ß can be subcloned.
Protocol using the QuickChange Ligthning Kit from Stratagene:
https://2010.igem.org/Image:Freiburg10_SDM_PstI_TK30_05_10.tif
The follwoing PCR was used:
- 95 °C 2'
- 95 °C 20"
- 60 °C 1'
- 68 °C 2'
- Goto 2 repeat 17
- 68 °C 5'
After the PCR program was conducted, I added 1 µL of DpnI provided with the Kit to each sample and incubated it for 10 minutes.
For transformation XL-10 Gold cells with 2µL beta-mercaptoethanol have been used and plated on agar plates containing chloramphenicol.
Cellculture: Bad news
Something happened to our virus-production cell line AAV293.
Nearly all AAV293 cells are dead.
The whole Loop-Insertion Transfection from 3.10 is lost, our actual culture flasks aswell. We dont know exactly the cause of this cellular mass mortality. Maybe someting is wrong with the new FCS-stocks.
Repetition of pCerulean_VP1up_NLS_DARPin
Investigators: Achim
- The Test Digestion (1.10.) of the last attempts clones didn't look convincing. We sent clone 1 for sequencing yesterday, but we didn't recieve any results yet. That's why I decided to repeat the experiment today, in case results are negative.
- Details: See first run from 28.9.
Prep. Gel:
Harvest of viral stocks
Investigators:
Number of viral stocks: 4
- mVenus standard stock 10 x cm R/C: P4682 plates => Harvest in 2 50 ml Falcons, four times freeze thaw centrifugation step with 2500g for 10 min
- TKGMK stock 10 x cm2 plates R/C: P468 => Harvest in 2 50 ml Falcons, four times freeze thaw centrifugation step with 2500g for 10 min
- viral particles with GOI (mVenus) missing HGH one 6well plate => Harvest in 1 50 ml Falcon, four times freeze thaw centrifugation step with 2500g for 10 min
- viral particles with GOI (mVenus) missing beta globin one 6well plate => Harvest in 1 50 ml Falcon, four times freeze thaw centrifugation step with 2500g for 10 min
- viral particles with R/C missing HSPG binding motif one 6well plate => Harvest in 1 50 ml Falcon, four times freeze thaw centrifugation step with 2500g for 10 min
- viral particles with new standard R/C (intact HSPG binding motif) one 6well plate => Harvest in 1 50 ml Falcon, four times freeze thaw centrifugation step with 2500g for 10 min
Motivation:
The two new standard vectors are from now on called Control Vectors, to get valide data. The TKGMK Stock will be used for the MTT-ASSAY.
Repetition of Cloning lITR_phTERT_beta-globin_mGMK_TK30 into pSB1C3_hGH_rITR
Investigator: Anna
Comment: Second approach of Cloning lITR_phTERT_beta-globin_mGMK_TK30 into pSB1C3_hGH_rITR, first one didn't work (see labday 01.10.).
Vector name:
- pSB1C3_hGH_rITR (P186)
c= 129,6 ng/µl
Insert name:
- pSB1C3_lITR_phTERT_beta-globin_mGMK_TK30 (P673)
c= 266,4 ng/µl
| components | volume for Pp728 /µl | volume of P673 /µl |
| DNA | 11,6 | 7,5 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme XbaI | 1 | - |
| Enzyme EcoRI | 1 | 1 |
| Enzyme SpeI | - | 1 |
| H2O | 2,4 | 6,5 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
0,4 g Agarose, 50 ml TAE (0,8%), 3 µl GELRED , at 115 Volt
Gel extraction:
Was performed according to the standard protocol.
| rITR_hgH | lITR_phTERT_betaglobin_mGMK_TK30 | |
| Expected size of fragment | 2690 bp | 3045 bp |
| DNA-concentration [ng/µl] | 37,4 | 19,81 |
T4 Ligation:
| volume of vector | 1,1 µl |
| volume of insert | 6,9 µl |
Transformation:
Was done following the standard protocol using BL21 cells.
Seeding HT1080 and AAV431 for transduction
- 3*6well plates HT1080
- 3*6well plates A431
for transduction with following viral stocks:
- mVenus Control Vector to quantify the other stocks
- mVenus without beta-globin
- mVenus without HGH
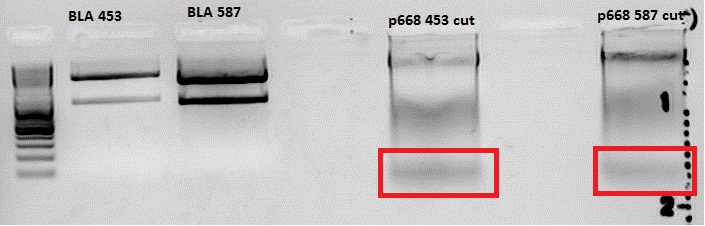
- mVenus in viral vectors without HSPG-Binding-site(P667)
- mVenus in viral vectors with HSPG-Binding-site, the new state-of-the-art-R/C (P668)
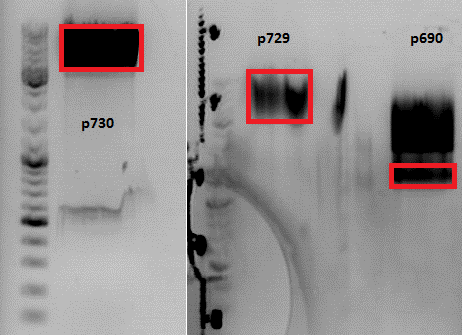
Cloning of pSB1C3_lITR_phTERT_beta-globin_CD and pSB1C3_lITR_CMV_beta-globin_CD
Investigator: Anissa
Vector name:
- pSB1C3_lITR_phTERT_beta-globin (p730)
- pSB1C3_lITR_CMV_beta-globin (P729)
- VB:pSB1C3_CD(P690)
Insert name:
| components | volume of P730 /µl | volume of P729 /µl | volume of P690 /µl |
| DNA | 12 | 4,1 | 9,9 |
| BSA (10x) | 2 | 1,5 | 2 |
| Buffer 4 (10x) | 2 | 1,5 | 2 |
| Enzyme AgeI | 1 | 1 | - |
| Enzyme NgoMIV | - | - | 1 |
| Enzyme PstI | 1 | 1 | 1 |
| H2O | 1 | 5,9 | 4,1 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 15 | 20 |
Gel:
for vectors and insert :
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 110 Volt
Gel extraction:
Was performed according to protocol.
T4 Ligation:
- Volume vector P729: 1,39 µl
- Volume insert: 6,61 µl
- Volume vector P730: 3,55 µl
- Volume insert: 4,45 µl
Transformation:
Was performed according to standard protocol using BL21 cells.
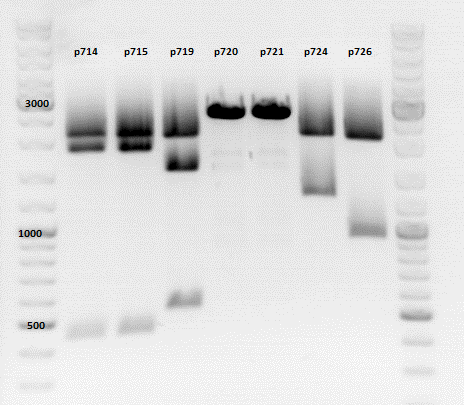
Repetition of the test-digestion of p714,p715,p719,p720,p721,p724 and p726
Investigator: Anissa
- pSB1C3_lITR_CMV_beta-globin_mCherry: Cut with EcoRI, PstI & NdeI
- P714 (Lane 1)
- P715 (Lane 2)
- Results:Looks good.
- pSB1C3_lITR_phTERT_beta-globin_mCherry: Cut with EcoRI, PstI & SacII
- P719 (Lane 3)
- Results:Looks good.
- P719 (Lane 3)
- pSB1C3_lITR_phTERT_beta-globin_CFP_hgh_rITR: Cut with EcoRI, AgeI & PstII
- P720 (Lane 4)
- P721 (Lane 5)
- Results:The expected bands should run at 671 bp,2033 bp and 1855 bp. Sequencing has to be repeated
- pSB1C3_lITR_phTERT_beta-globin_CD: Cut with PstI & EcoRI
- P724 (Lane 6)
- P726 (Lane 7)
- Results:The expected bands should run at 2035 and 2460 bp. The experiment has been repeated already today.
Repetition: Cloning of VP2/3_BAP_HSPG-KO and VP2/3_His_HSPG-KO into pCerulean_Zegfr:1907_Middlelinker and VP2/3_His_HSPG-KO into pCerulean_VP1up_NLS_mVenus
Investigator: Stefan
Comment: Cloning of 587-ko_His and 587-ko_BAP into pSB1C3_001_VP2/3_capins did not work out last time. Since cloning of VP2/3_capins started before sequencing results arrived, this experiment has to be repeated.
Vector name:
- pCerulean_Zegfr:1907_Middlelinker (P408)
- pCerulean_VP1up_NLS_mVenus (P426)
Insert name:
- pSB1C3_001_VP2/3_capins_587_KO_His_clone3 (P734)
- pSB1C3_001_VP2/3_capins_587_KO_Bap_clone1 (P738)
| components | volume for inserts (P734 + P738) /µl | volume of P408 /µl | volume of P426 /µl |
| DNA | 14 | 3 | 3 |
| BSA (10x) | 3 | 2 | 2 |
| Buffer 4 (10x) | 3 | 2 | 2 |
| Enzyme NgoMIV | 1 | - | - |
| Enzyme PstI | 1 | 1 | 1 |
| Enzyme MscI | 1 | - | - |
| Enzyme AgeI | - | 1 | 1 |
| H2O | 7 | 11 | 11 |
| Total volume (e.g. 15,20,25,30 µl) | 30 | 20 | 20 |
Gel:
0,4 g Agarose, 50 ml TAE (0,8%), 3 µl GELRED , at 115 Volt
Gel extraction:
Was performed according to protocol.
T4 Ligation:
| ligation name | 408 + 734 | 408 + 738 | 426 + 734 | 426 + 738 |
| volume of vector | 3,81 | 3,81 | 3,56 | 3,56 |
| volume of insert | 4,19 | 4,19 | 4,44 | 4,44 |
Transformation:
Was performed according to standard protocol using BL21 cells.
PCR of VP1-3
Investigator: Kira
Comment: this approach was already repeated at least 2 times, but still even if its not one of the very important constructs, it should be finished by end of the month.
PCR*PCR program:
c(pKEX_VP1)=488 ng/ul
c(pKEX_VP2)=484 ng/ul
c(pKEX_VP2)=485 ng/ul
VP-1 praefix primer: 089 VP-2 praefix primer: 090 VP-3 praefix primer: 091 VP1-3 suffix reg_25: 0120
| components | volume in µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| primer_for (1:10 dilution) | 2,5 |
| primer_rev (1:10 dilution) | 2,5 |
| DNA template (1:100) | 0,5 |
| DMSO | 0 |
| Phusion polymerase | 0,5 |
| H2O | 33 |
| Total volume (e.g. 50 µl) | 50 |
The PCR programm for all 3 samples was the same due to the weakest primer, which is VP-1 praefix primer
| Cycles | Temperature | Time |
| 98°C | 30 sec | |
| 10x | 98°C | 10 sec |
| 60°C | 25 sec | |
| 72°C | 1 min 6 sec | |
| 20x | 98°C | 15 sec |
| 72°C | 1 min 31 sec | |
| 1x | 72°C | 5 min |
| Hold 4°C |
1% agarose gel
The gel shows only the primer bands on the bottom of the gel --> PCR failed again. Its going to be repeated tomorrow.
141. labday 06.10.2010
Cellculture
Investigator Patrick
Not only the 293 cells look bad. The A431 cells and the HT1080 cells die, too. There is no contamination detectable. Maybe there is something wrong with the DMEM because the now 100 % serum-free AAV293 cells dont die.
Transduction of HT1080 and A431 cells
The cells are not adherent so no transduction was performed.
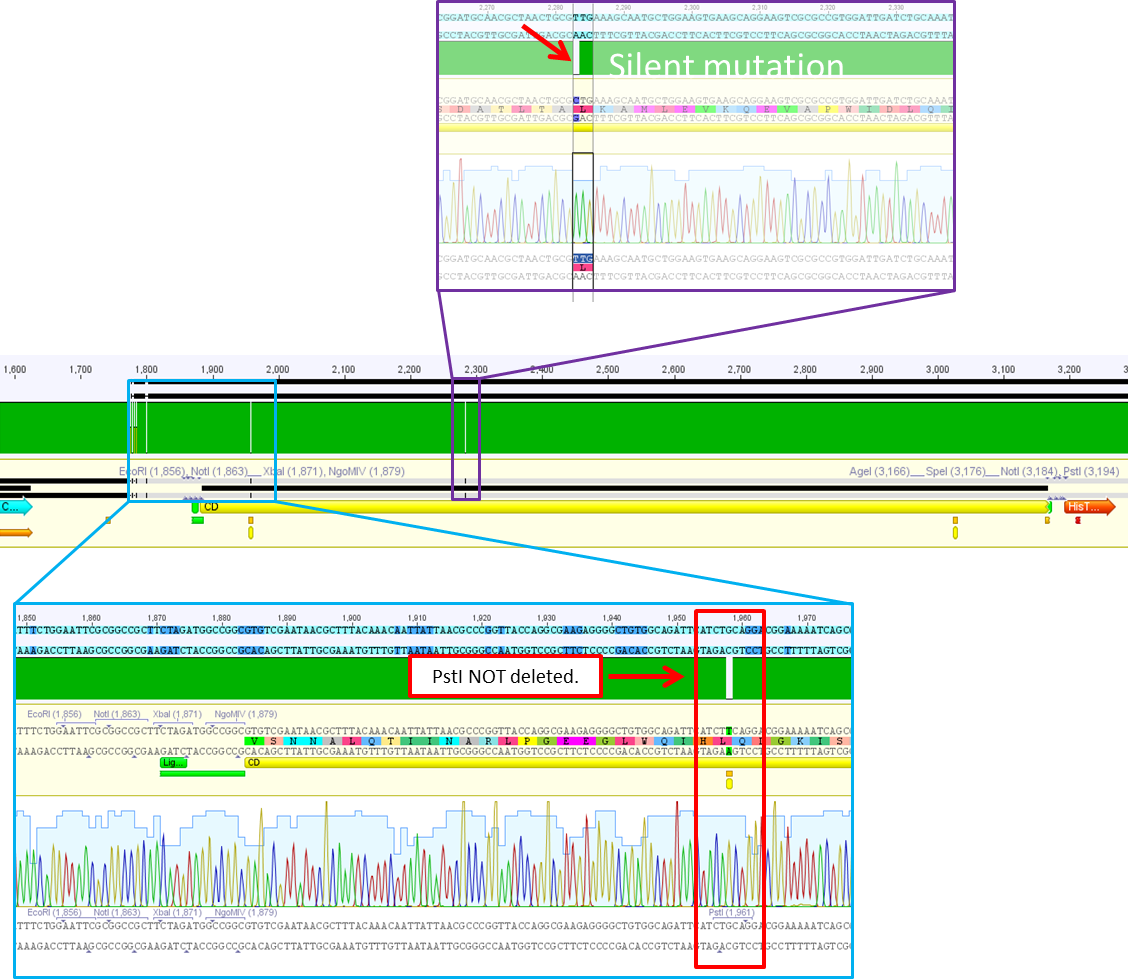
Sequence analysis of pSB1C3_CD
Investigator: Bea
Comment: After test digestion revealed positive results, we sent one clone for sequencing (P690) with VF-2 and VR-2 primers. The tube numbers are: SB_1 and SB_2.
Results:
- Prefix looks well. Alle restriction sites can be found in the prefix RFC25.
- Suffixlooks well. Alle restriction sites can be found in the suffix RFC25.
- BUT: There seems to be a PstI recognition site within the sequence. Therefore the site-directed mutagenesis needs to be repeated.
Site-directed mutagenesis of pSB1C3_CD
Investigator: Bea
Comment: Since the sequence analysis revealed that the PstI restriction iste within the CD is NOT removed I prepared another Quickchange in order to mutate the PstI site.
Protocol:
The following PCR was used:
- 95 °C 2'
- 95 °C 20"
- 60 °C 10"
- 68 °C 1'45"
- Goto 2 repeat 17
- 68 °C 5'
After the PCR program was conducted, I added 1 µL of DpnI provided with the Kit to each sample and incubated it for 10 minutes at 37°C.
For transformation XL-10 Gold cells with 2µL beta-mercaptoethanol have been used and plated on agar plates containing chloramphenicol.
Sequencing analysis
Investigator: Stefan
Comment:
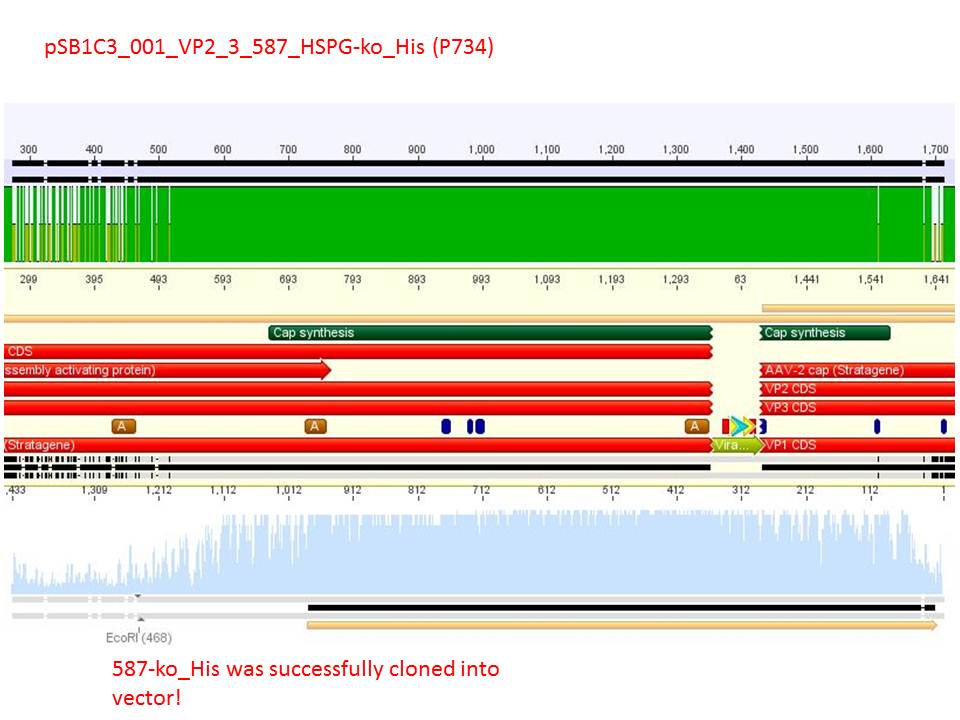
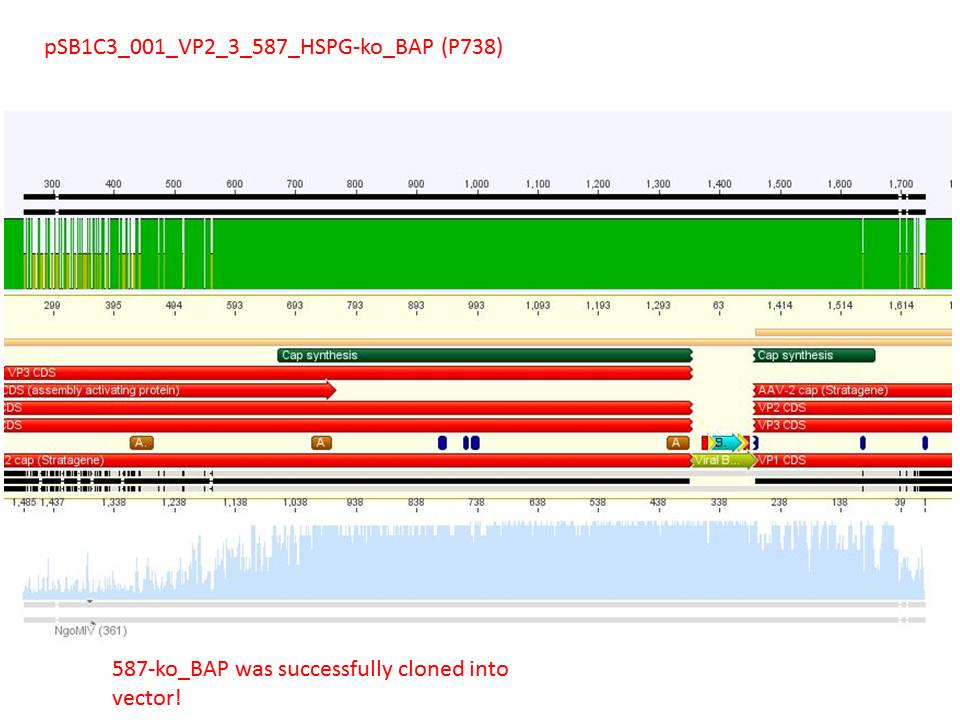
Sequencing reults of pSB1C3_CMV_ZEGFR:1907_VP2/3_CapIns (P679), pSB1C3_CMV_ZEGFR:1907_VP2/3_CapIns_HSPG-ko (P684), pSB1C3_001_VP2/3_587_HSPG-ko_His(P734) and pSB1C3_001_VP2/3_587_HSPG-ko_BAP(P738). In P679 and P684, CMV_ZEGFR:1907 was cloned in front of VP2/3, in P734 and P738, 587_HSPG-ko_His/BAP was cloned into the 587 loop.
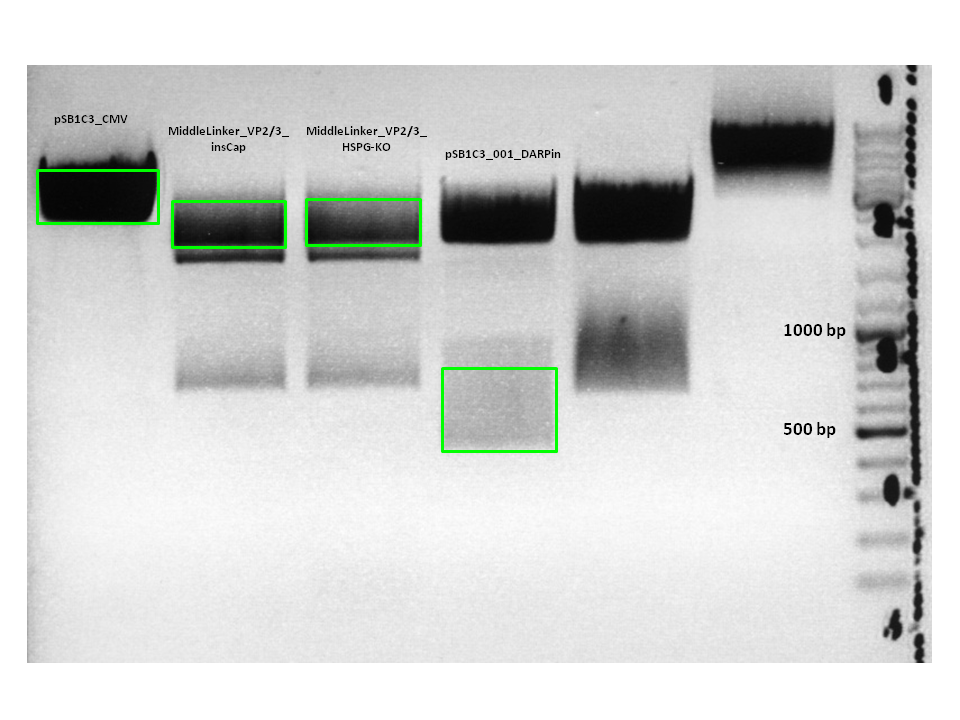
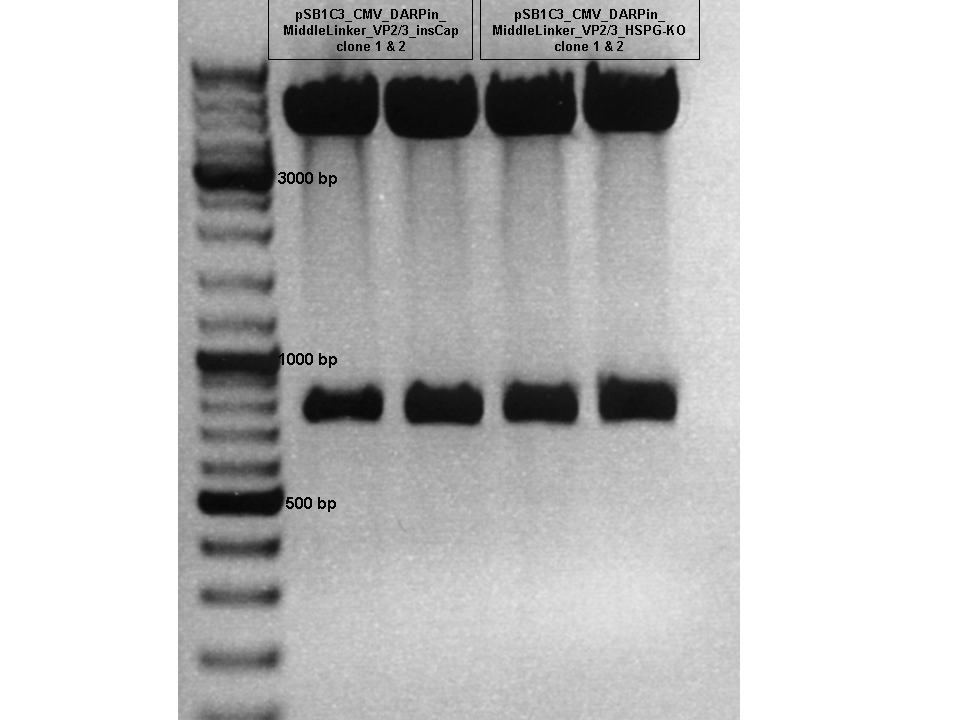
Test Digestion of pSB1C3_CMV_DARPin_MiddleLinker_VP2/3_insCap and pSB1C3_CMV_DARPin_MiddleLinker_VP2/3_HSPG-KO
Investigator: Hanna
Comment: Besides the Affibody we also want to test and characterize the DARPin E01 as a targeting motif for EGFR over expressing tumor cells. The day before yesterday a 3 fragment ligation was performed with the CMV promoter, DARPin and pSB1C3_VP2/3_insCap/pSB1C3_VP2/3_HSPG-KO.
2 clones were picked from each approach and preped today.
TEST DIGESTION:
- DNA: 2 µL
- Buffer 3: 1 µL
- BSA (10x): 1 µL
- XbaI: 0.5 µL
- BmgBI (cuts in the DARPin sequence): 0.5 µL
- H2O: 5 µL
Incubation: 2 hours.
Conclusion: Test digestion looks well: Expected fragments = 756 bp and 4409 bp.
Two samples were sent for sequencing to GATC using the CMV-F primer.
Repetition: Cloning of CFP into pSB1C3_lITR_phTERT_beta-globin and hgH_rITR into pSB1C3_lITR_CMV_beta-globin_mCherry and into pSB1C3_lITR_phTERT_beta-globin_mCherry
Investigator: Stefan
Comment:
Vector name:
- pSB1C3_lITR_phTERT_beta-globin (P437)
- pSB1C3_lITR_CMV_beta-globin_mCherry (P715)
- pSB1C3_lITR_phTERT_beta-globin_mCherry (P719)
Insert name:
- pSB1C3_CFP (P666)
- pSB1C3_hgH_rITR (P728)
| components | volume for inserts (P666 + P728) /µl | volume of vectors P437 + P715 + P719 /µl |
| DNA | 10 | 4 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme PstI | 1 | 1 |
| Enzyme XbaI | 1 | - |
| Enzyme SpeI | - | 1 |
| H2O | 4 | 10 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
Gel:
0,4 g Agarose, 50 ml TAE (0,8%), 3 µl GELRED , at 115 Volt
Gel extraction:
Was performed according to protocol.
T4 Ligation:
| ligation name | 437 + 666 | 715 + 728 | 719 + 728 |
| volume of vector | 5,08 | 5,13 | 5,34 |
| volume of insert | 2,92 | 2,87 | 2,66 |
Transformation:
Was performed according to standard protocol using BL21 cells.
Biotinylation of the monoclonal IgG antibody A20
The biotinylation of the A20 antibody that recognizes the assembled virus was performed with a biotinylation Kit purchased from Dojindo.
- The first biotinylation reaction was performed according to the protocol with 150µg of the purified antibody but resulted in not detectable protein concentrations.
- The biotinylation reaction was repeated for a second time with 200µg and resulted in satisfying protein concentrations.
- Successful biotinylation has to be tested in an other experiment.
Repetition of VP1-3 PCR
Investigator: Kira
PCR program:
c(pKEX_VP1)=488 ng/ul
c(pKEX_VP2)=484 ng/ul
c(pKEX_VP2)=485 ng/ul
VP-1 praefix primer: 089 VP-2 praefix primer: 090 VP-3 praefix primer: 091 VP1-3 suffix reg_25: 0120
Regarding the primer sequences, DMSO will be added to the PCR samples.
| components | volume in µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| primer_for (1:10 dilution) | 2,5 |
| primer_rev (1:10 dilution) | 2,5 |
| DNA template (1:100) | 0,5 |
| DMSO | 2 |
| Phusion polymerase | 0,5 |
| H2O | 31 |
| Total volume (e.g. 50 µl) | 50 |
The PCR programm for all 3 samples was the same due to the weakest primer, which is VP-1 praefix primer
| Cycles | Temperature | Time |
| 98°C | 30 sec | |
| 10x | 98°C | 15 sec |
| 59°C | 25 sec | |
| 72°C | 1 min 11 sec | |
| 20x | 98°C | 15 sec |
| 72°C | 1 min 36 sec | |
| 1x | 72°C | 5 min |
| Hold 4°C |
1% agarose gel
File:Freiburg10 VP1-3 PCR.jpg
Digestion of plasmid backbone:
pSB1C3_001 is used as backbone
| Components | vector Volume/µL |
| DNA | 3,0 µl |
| BSA (10x) | 2 µl |
| Buffer no. 4 (10x) | 2,0 µl |
| Enzyme 1 AgeI | 1,5 µl |
| Enzyme 2 SpeI | 1,0 µl |
| H2O | 10,5 µl |
| Total volume | 20 |
incubation @ 37 C for approx. 2 h
1% agarose gel
File:Freiburg10 digestion-pSB1C3 001.jpg
Digestion of PCR product:
| Components | PCR product Volume/µL |
| DNA | 30,0 µl |
| BSA (100x) | 4,5 µl |
| Buffer no. 4 | 4,5 µl |
| Enzyme 1 NgoMIV | 1 µl |
| Enzyme 2 SpeI | 1,0 µl |
| H2O | 4 µl |
| Total volume | 45 |
incubation @ 37 C for approx. 2 h
ligation with T4 ligase was performed over-night
Cloning of Rep into Rep52 and Rep78
Investigator: Kira
Comment: In order to perfom PCR, we have to clone the ordered Rep gene into Rep52 and Rep78 backbones.
Regarding the activity conditions of the enzymes, digestion was performed in 2 steps.
| Components | insert | Rep78 | Rep52 |
| DNA | 3 µl | 2 µl | 7 µl |
| BSA (100x) | 2 µl | 2 µl | 2 µl |
| Buffer no. 3 | 2,0 µl | 2,0 µl | 2,0 µl |
| Enzyme SwaI | 1,0 µl | 1,0 µl | 1,0 µl |
| H2O | 12 µl | 13 µl | 8 µl |
| Total volume | 20 |
incubation @ 25 C for approx. 2 h
Dephosphorylation was performed because of the blunt cleavage. PCR phosphorylation followed in order to get rid off the buffer because Buffer 3 is not appropriate for further digestion.
after PCR phosphorylation:
c(pMA_RC-insert)= 21,75 ng/ul
c(rep52)= 75,82 ng/ul
c(rep78)= 2,47 ng/ul
| Components | insert | Rep78 | Rep52 |
| DNA | 30 µl | 30 µl | 30 µl |
| BSA (100x) | 0 µl | 0 µl | 0 µl |
| Buffer no. 2 | 4,5 µl | 4,5 µl | 4,5 µl |
| Enzyme HindIII | 1,0 µl | 1,0 µl | 1,0 µl |
| H2O | 9,5 µl | 9,5 µl | 9,5 µl |
| Total volume | 45 |
incubation for 2h at 37C
the gel shows that Rep78 backbone got lost.. Regarding the concentration, which was measured after PCR purification it might be possible --> digestion of Rep78 will be repeated tomorrow, while Rep52 is ready for ligation.
142. labday 07.10.2010
DARPin VP2 Fusion: Sequencing results
Investigator: Hanna
COMMENT: The DARPin was fused with the Middle Linker to the VP2/3 BioBrick and cloned into the pSB1C3 backbone. Yesterday's test digestion looked well, so one clone of the VP2/3_insCap and one clone of the VP2/3_HSPG-KO construct was sent for sequencing.
CONCLUSION: Sequencing looked well: 3 fragment ligation was successful - because the CMV-F primer was used for sequencing, the CMV promoter was successfully inserted. As shown in the alignments also the DARPin was inserted. This is the first time that the DARPin was sequenced - thus the original sequence must also be OK!
These two constructs are finished and are ready to be tested in cell culture via qPCR, ELISA,... and can be stored in our BioBrick Box for submitting.
Repetition of cloning of ordered Rep into Rep78
Investigator: Kira
Comment: Because the gel from yesterday did not show any bands in the Rep78 line, I desided to repeat this approach but to use more DNA, in order to be able to continue with the second digestion (see labday 06.10.2010)
The same procedure but with 7 ul Rep78 (c=556 ng/ul). After the first digestion, dephosporylation and PCR purification were performed and DNA was measured.
As a control, the ordered Rep was used again.
c(p190)= 28 ng/ul
c(Rep78)= 6,90 ng/ul
the second digestion as performed despite the low concentration.
File:Freiburg10 2010-10-07.jpg
The gel shows once again no detectable DNA of Rep78.
Transformation of VP1-3 & pSB1C3_001 and p190 &Rep52
Investigator: Kira
The transformation was performed according to the standard protocol with BL21. Compared to VP1-3 transformed cells, Rep52 cells revealed no visible pellet after centrifugation, thus 300 ul of the cells suspension was plated.
Test digestion of different constructs
Investigator: Anna
Gel1_Part1: The 453 constructs were cut with BamHI and SspI, the 587 constructs with SalI and PvuII.
The inserts were cloned into pSB1C3_RepCap_p5tatless.
All of the following constructs were cut with EcoRI and PstI:
143. labday 08.10.2010
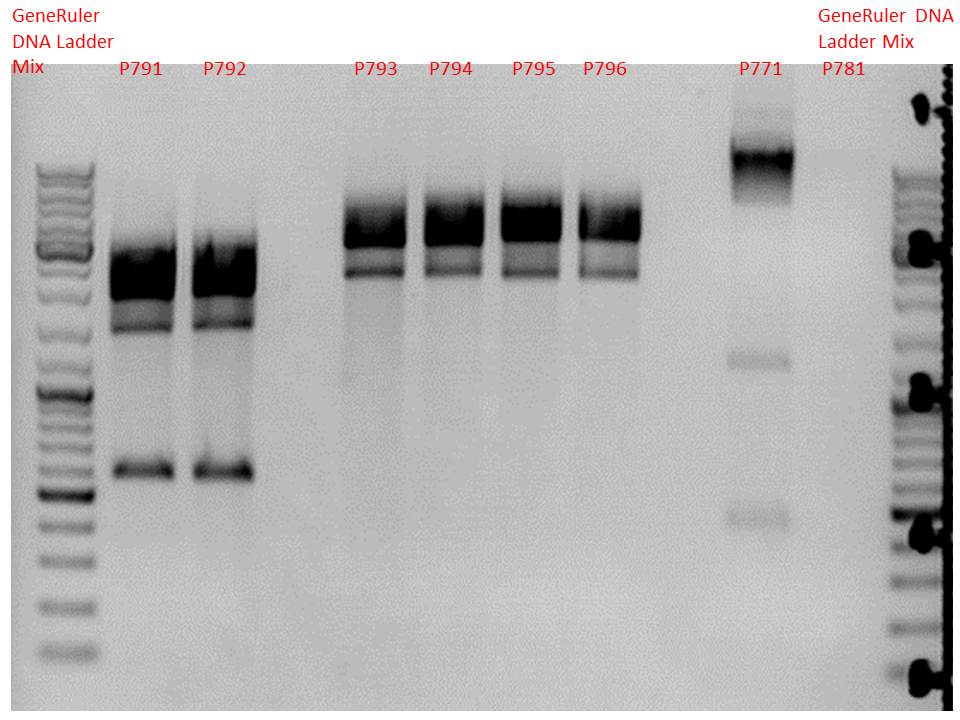
Mini-Prep and test digestion of several constructs
Investigator: Stefan
Comment: Mini-Preps were performed according to protocol:
Glycerol stocks were prepared:
- B641 pSB1C3_lITR_phTERT_beta-globin_CFP clone 1
- B642 pSB1C3_lITR_phTERT_beta-globin_CFP clone 2
- B643 pSB1C3_lITR_CMV_beta-globin_mCherry_hgH_rITR clone 1
- B644 pSB1C3_lITR_CMV_beta-globin_mCherry_hgH_rITR clone 2
- B645 SB1C3_lITR_phTERT_beta-globin_mCherry_hgH_rITR clone 1
- B646 pSB1C3_lITR_phTERT_beta-globin_mCherry_hgH_rITR clone 2
Mini-Prep was performed according to standard protocol:
- P791 pSB1C3_lITR_phTERT_beta-globin_CFP clone 1 c = 244.3 ng/µl
- P792 pSB1C3_lITR_phTERT_beta-globin_CFP clone 2 c = 214.3 ng/µl
- P793 pSB1C3_lITR_CMV_beta-globin_mCherry_hgH_rITR clone 1 c = 209.2 ng/µl
- P794 pSB1C3_lITR_CMV_beta-globin_mCherry_hgH_rITR clone 2 c = 262.4 ng/µl
- P795 pSB1C3_lITR_phTERT_beta-globin_mCherry_hgH_rITR clone 1 c = 234.7 ng/µl
- P796 pSB1C3_lITR_phTERT_beta-globin_mCherry_hgH_rITR clone 2 c = 207.2 ng/µl
Test digestion:
Comment:Additionally, P771 and P781 from yesterday's Mini-Prep were digested again.
| Components | P791 + P792 / µl | P793 - P796 / µl | P771 / µl | P781 / µl |
| DNA | 1,5 | 1 | 3 | 7,7 |
| Buffer 4 | 1 | 1 | 1 | 1 |
| BSA (10x) | 1 | 1 | 1 | 1 |
| PstI | 0,3 | 0,3 | 0,3 | 0,3 |
| EcoRI | 0,3 | 0,3 | - | - |
| BpmI | 0,3 | - | - | - |
| H2O | 5,6 | 6,4 | 4,2 | - |
| Total volume | 10 | 10 | 10 | 10 |
Comment:Samples P791 to P796 perform like expected, they can be used for further cloning or testing in cell culture. P771 show strange bands, a test PCR will be performed to verify successful cloning. P781 does not show any DNA at all like last time. A new SDm will be performed for pSB1C3_mGMK_TK30 (P675).
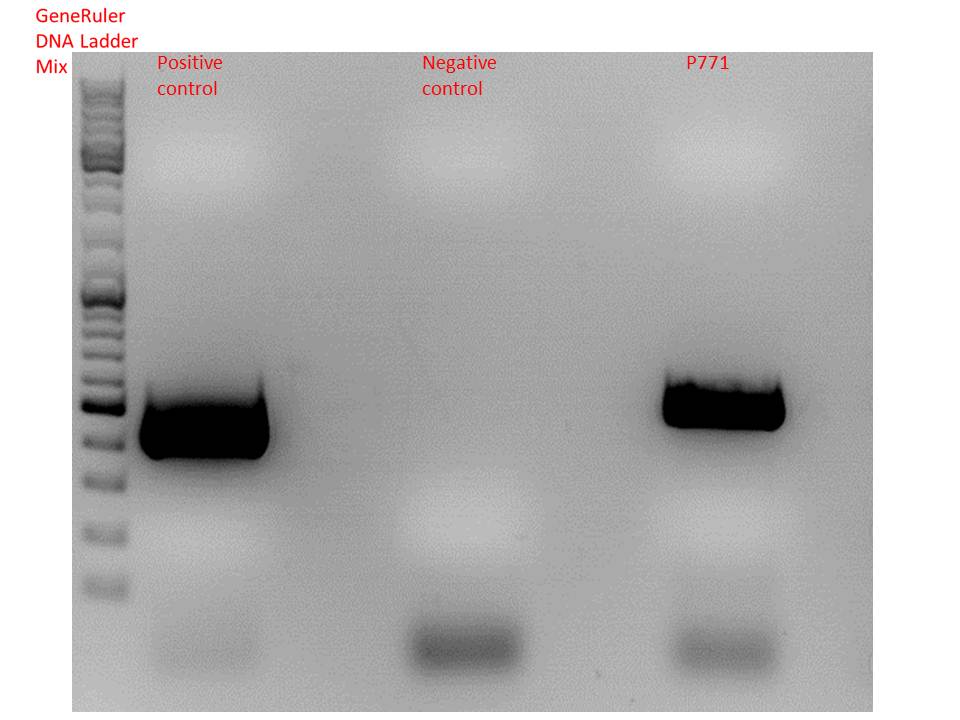
Test-PCR of pSB1C3_lITR_phTERT_beta-globin_mGMK_TK30_hgH_rITR
Investigator: Stefan
Comment: Test digestion showed strange results (an additional band at 500bp which does not fit any expectations), therfore an additional test-PCR was performed to verify successful cloning of the complete constructs.
| Components | positive control / µl | negative control / µl | sample P771 / µl |
| 5x Phusion HF buffer | 10 | 10 | 10 |
| 10mM dNTP mix | 1 | 1 | 1 |
| Primer for (1:10 dilution) | O107 2,5 | O29 2,5 | O29 2,5 |
| Primer rev (1:10 dilution) | O108 2,5 | O191 2,5 | O191 2,5 |
| DNA template | 1 (1:100) | 1 (1:100) | 1 (1:100) |
| DMSO | 1 | 1 | 1 |
| Phusion Polymerase | 0,5 | 0,5 | 0,5 |
| H2O | 32,5 | 32,5 | 32,5 |
| Total volume | 50 | 50 | 50 |
Gel:
0,5 g agarose, 50 ml TAE (1%), 115V, 50 minutes
Comment: Test PCR showed successful cloning of lITR_phTERT_beta-globin_mGMK_TK30 into pSB1C3_hgH_rITR. SDM to remove PstI in TK30 can be performed.
Trafo evaluation of VP1-3 and Rep52 constructs
Investigator: Kira
VP1-3 contain lots of colonies, while Rep52 plate is empty
Repetiton: Site-directed mutagenesis in pSB1C3_mGMK_TK30 construct
Investigator: Stefan
Comment: The first try to remove the PstI restriction site delivered no plasmid DNA, so another approach needs to be started.
Protocol using the QuickChange Lightning Kit from Stratagene:
Ingredients:
| Ingredients | Volume / µl |
| 10x reaction buffer | 2,5 |
| dNTP | 0,5 |
| forward primer: O191 | 0,69 |
| reverse primer: O192 | 0,71 |
| DNA Template | 5 |
| QuikSolution Reagent | 0,75 |
| QuikChangeLightning Enzyme | 0,5 |
| H2O | 14,35 |
| Total volume | 25 |
The following PCR program was used:
- 95 °C 2 minutes
- 95 °C 20 seconds
- 60 °C 1 minute
- 68 °C 2 minutes
- Goto step 2 repeat 17 times
- 68 °C 5 minutes
Digestion using DpnI:
To digest methylated and hemi-methylated DNA, 1 µl of DpnI was added and the mixture was incubated for 10 minutes at 37 °C.
Transformation:
Since XL-10 Gold cells delivered a bacteria lawn on the plates, this time XL1b cells were used.
144. labday 09.10.2010
Mini-Prep of pSB1C3_CD_SDM-PstI
Investigator: Stefan
Comment: Mini-Prep was performed according to standard protocol.
Glycerol stocks were prepared:
- B647 = pSB1C3_CD_SDM-PstI clone 1
- B648 = pSB1C3_CD_SDM-PstI clone 2
Mini-Prep was performed according to standard protocol:
- P797 = pSB1C3_CD_SDM-PstI clone 1 c = 8,2 ng/µl
- P798 = pSB1C3_CD_SDM-PstI clone 2 c = 7,1 ng/µl
Comment: Since Mini-Prep from XL-10 cells yielded that little plasmid, a re-trafo will be performed using BL21 cells.
Transformation:
A 1:10 dilution of both plasmids was prepared and 1 µl was added to BL21 cells.
Transformation was performed according to standard protocol. Cells were plated on agar plates containing chlorampenicol.
Preparation of competent cells and Test-Trafo
Investigator: Stefan
Preparation of BL21 (red line) and XL1-blue (green X) cells was performed according to standard protocol.
Do not use cells until further notice!
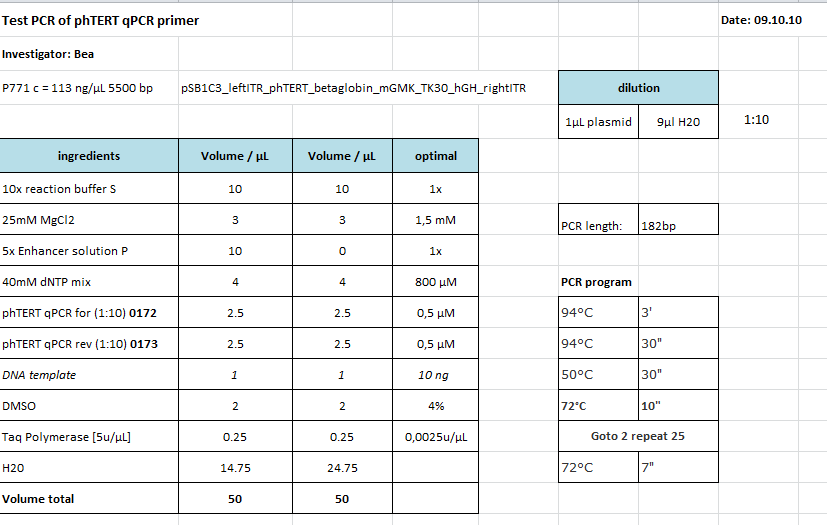
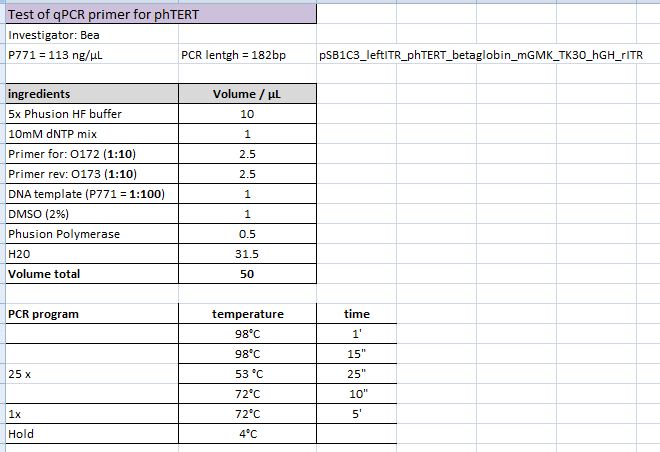
Test PCR of qPCR primer for phTERT
Investigator: Bea
Comment: The GOI which is flanked by the ITRs and encapsidated in the capsid is the mGMK_T30 driven by the pHTERT promoter which is only active in several tumor cells. Since we have never tested the primers if they work I wanted to do that. This time I tried to use the TAQ polymerase which we received from Peqlab. Since I could not find a standard protocol for the PCR provided by peqlab I tried to compare several manuals and combine them.
Protocol:
I did not tried the approach with ENhancer Solution.
I checked the protocol today (10.10) and recognized that I added accidentially to much buffer to the PCR-Mix. Maybe this could be another explanation for the negative results of the PCR.
The following PCR was used:
- 94 °C 3'
- 94 °C 30"
- 50 °C 30"
- 72 °C 10"
- Goto 2 repeat 17
- 68 °C 7'
Result:
Unfortunately the expected PCR product with a size of 182 bp could not be detected as it can be seen in the agarose gel. I can not say if the protocol needs to be modified or if the primer pair did not anneal adequatly. Therefore, I am going to repeat the PCR tomorrow by using the Phsuion polymerase for checking the results.
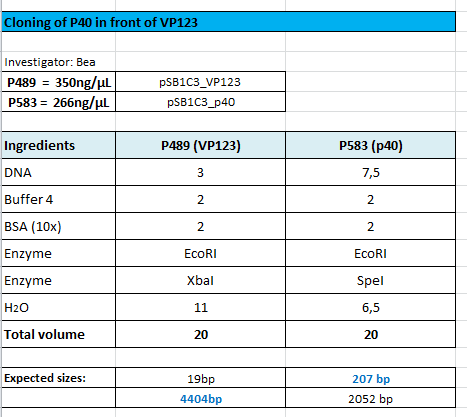
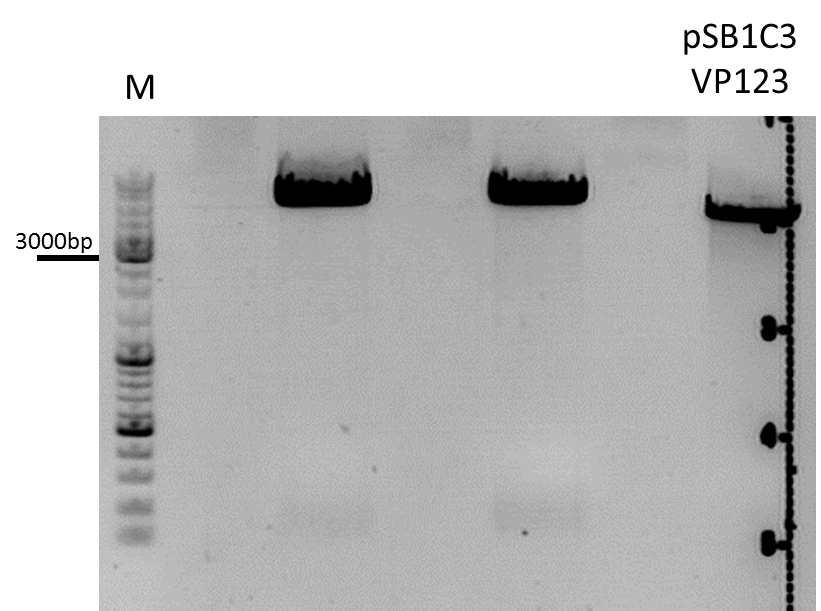
Cloning of p40 in front of VP123
Investigator: Bea
Comment: We want to test the promoter strenth of several promoters (CMV, P40, p5, p5tataless) and compare them in order to provide the possibility of choosing the proper promoter for each experiment and to titrate the plasmids during transfection. Therefore, the p40 promoter needs to be fused in front of the VP123 construct in order to compare the CMV and the p40 promter
Protocol:
The following results of the agarose gel can be seen. The fragments were cut out and a gel extraction has been performed:
For transformation BL21 cells have been used and plated on agar plates containing chloramphenicol.
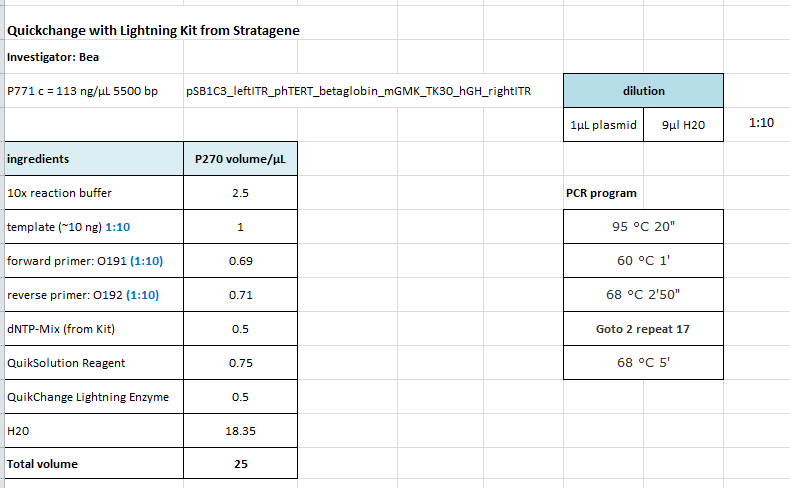
Site-directed mutagenesis of pSB1C3_leftITR_phtert_betaglobin_mgmktk30_hGH_rITR
Investigator: Bea
Comment: Since we still have the PstI restriction site within the sequence we need to delete the restriction enzyme site.
Protocol:
The following PCR was used:
- 95 °C 2'
- 95 °C 20"
- 60 °C 10"
- 68 °C 2'50"
- Goto 2 repeat 17
- 68 °C 5'
After the PCR program was conducted, I added 1 µL of DpnI provided with the Kit to each sample and incubated it for 10 minutes at 37°C.
For transformation XL-1 cells have been used because the bacterias of the last site-directed mutageneis approaches using XL-10 cells showes extensive grwoth (bacterial lawn). I plated on the transformaned cells on agar plates containing chloramphenicol.
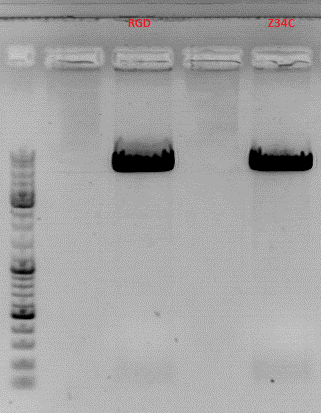
Cloning HSPG-KO_empty into pSB1C3-001_pCMV_[AAV2]VP123_453_RGD and pSB1C3-001_pCMV_[AAV2]VP123_453_Z34C
Investigator: Achim
- In order to obtain transduction data for our targeting approaches, we still need constructs with RGD/Z34C in the 453 loop as well as HSPG_KO_empty in the 587 loop.
- I digested pSB1C3_HSPG-KO_empty (P611) as well as pSB1C3-001_pCMV_[AAV2]VP123_453_RGD (P634) and pSB1C3-001_pCMV_[AAV2]VP123_453_Z34C (P641)with BamHI and PvuII and ligated the HSPG-KO_empty motive into both vectors.
- Digestion:20µl,Buffer 4, BSA, 1µl enzyme, 37°
- VB:pSB1C3_001_587_KO_empty (P611)
- c=160 ng/µl ->14 µl
- pSB1C3-001_pCMV_[AAV2]VP123_453_RGD (P634)
- c=239 ng/µl (1:10 dilution of midiprep)->5 µl
- pSB1C3-001_pCMV_[AAV2]VP123_453_Z34C (P641)
- c=229 ng/µl (1:10 dilution of midiprep) -> 5µl
- VB:pSB1C3_001_587_KO_empty (P611)
- Dephosphorylation: 1h, 37°C, antarctic phosphatase
- Prep gel:
- Ligation: T4
Cellculture
Investigator Patrick
- mVenus without beta-globin
- mVenus without HGH
- mVenus HSPG KO
- mVenus standard
- mVenus new standard
Expected results:
- mVenus without beta-globin: beta globin is said to improve the expression therefore we expect a worse mVenus expression compared to the standard AAVs.
- mVenus without HGH: HGH is responsible for the mRNA polyadenylation of the virus so we expect expect a worse mVenus expression compared to the standard AAVs, too.
- mVenus HSPG KO: the primary AAV2 receptor (the HSPG receptor) was knocked out therefore we expect a worse tranduction rate (about 0,3 to 0,4 of the standard transduction rate, assuming that there are HSPGs on the HT1080 and A431 cell surface). The worse the better.
- mVenus standard: this will be the reference for the other four transduction experiments (no point mutations, GOI: mVenus)
- mVenus new standard: AAV2 with 4 point mutations, inserted rep & cap and "remutated" KpnI restriction site hopefully yiels results similar to the mVenus standard.
145. labday 10.10.2010
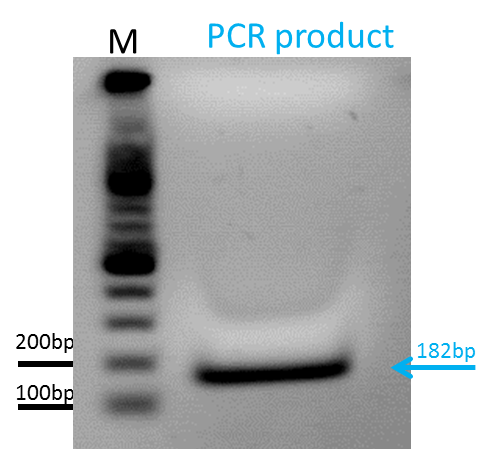
Repetition of Test-PCR of qPCR primer for phTERT
Investigator: Bea
Comment: The GOI which is flanked by the ITRs and encapsidated in the capsid is the mGMK_T30 driven by the pHTERT promoter which is only active in several tumor cells. Since we have never tested the primers if they work I wanted to do that. For several reason I decided to use the Phusion polymerase to perform the PCR. First, we are familiar with the handling of this polymerase and have a protocol which we already used several times and second, since I could not find any standard protocols for the TAQ-polymerase provided by PeqLab, as mentioned yesterday, I will rely on the Phusion polymerase.
Protocol:
Results: After PCR was completed, I loaded a 1.5% agarose gel. The results can be seen in the picture: There is a intensive band aat around 150-180bp which correspond to the expected fragment size of 182 bp. We can conclude that the the primers work and that they can be used in the qPCR approaches. One note along the way: I´ve missed the fact that the buffer in which the gel was loaded, was TBE and not TAE. Still the fragment looks well, even though the gel did not appeared to ran normal.
New XL1b and BL21 cells ready to use!
Investigator: Stefan
Cloning Super Constructs and VP2 N-terminal Fusion_ HSPG-KO constructs into pSB1C3_001_CMV or pSB1C3_CMV
Investigator: Hanna
Comment: In order to send our final VP2 N-terminal Fusion constructs with HSPG konock down and our super constructs to the registry, they need to be cloned into the pSB1C3 backbone.
We would prefer sending them in the pSB1C3_001 plasmid, because it does its ViralBricks restriction sites were deleted. Therefore one can cut out the 587_KO ViralBrick and replace it by any ViralBrick of interest.
Because test digestion of pSB1C3_001_CMV didn't show any clear results, I will digest both clones and in addition to that pSB1C3_CMV.
Incubation: 2 h, 37°C
Midi-Prep
Investigator: Chris W.
Midi-Prep of:
pCerulean_VP1up_NLS_Zegfr:1907_VP2/3-Capins clone 2 =P799 =B401
pSB1C3_CMV_DARPin_MiddleLinker_VP2/3_insCap clone 2 =P800 =B595
pSB1C3_CMV_DARPin_MiddleLinker_VP2/3_HSPG-KO clone 1 =P801 =B596
pSB1C3_left-ITR_phTERT_beta-globin_mGMK_TK30_hGH_right-ITR clone 1 =P802 =B622
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P799 | P800 | P801 | P802 |
| concentration (ng/µl) | 662,8 | 911,3 | 495,6 | 673,9 |
Checking trafo plates of Viralbrick approaches, SDM of pSB1C3_leftITR_phtert_betaglobin_mgmktk30_hGH_rITR and P40_VP123
Investigator: Bea
Comment: Unfortunately, no cells grew on the agar plates containing cm. Therefore, we repeat the Trafo ones again and plate them on new agar plates hoping that colonies will grow over night.
I used the ligations prepared yesterday and transformed new bacterial cells (XL-1B for SDM and BL21 for the other three ligations):
- pSB1C3_p40_VP123
- P771_SDM (PstI): pSB1C3_leftITR_phtert_betaglobin_mgmktk30_hGH_rITR
- pSB1C3-001_pCMV_[AAV2]VP123_453_RGD_587-ko
- pSB1C3-001_pCMV_[AAV2]VP123_453_Z34C_587-ko
Preparations for ELISA
Investigator: Volker M.
Two 96-well plates that were coated with 200µg A20 antibody per well for 48h at 4°C were washed three times with 200µl PBST Buffer and incubated over night at 4°C with 300µl PBST + 0.2 % BSA.
As a preparation 4 liters of PBS buffer were prepared.
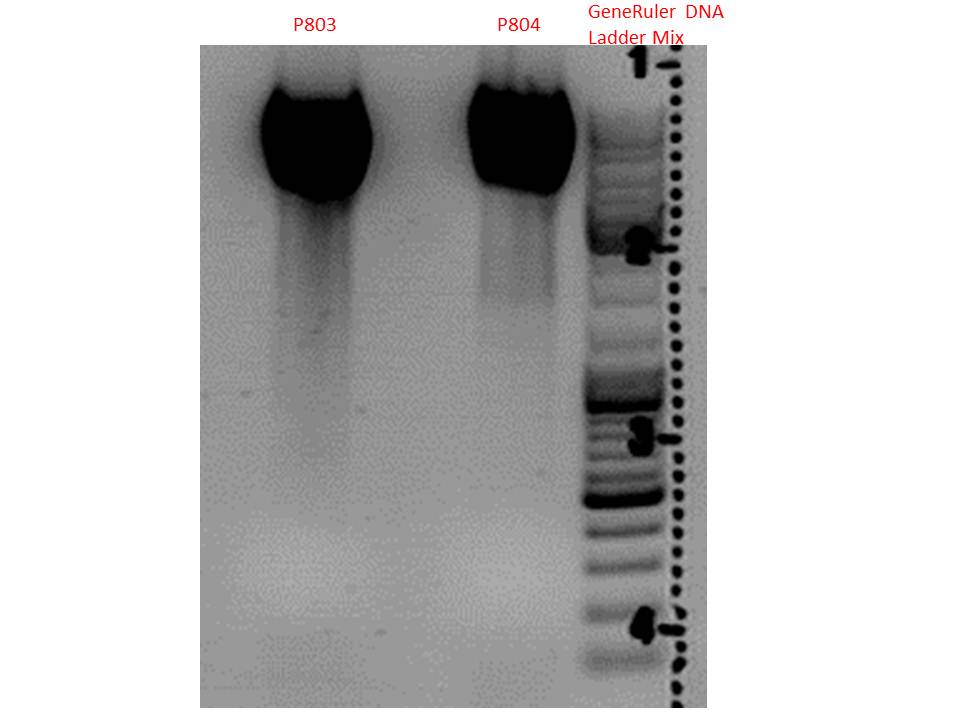
Mini-Prep of pSB1C3_gMK_TK30_SDM-PstI_new
Investigator: Stefan
Glycerol stocks were prepared:
- B649 = pSB1C3_gMK_TK30_SDM-PstI_new clone 1
- B650 = pSB1C3_gMK_TK30_SDM-PstI_new clone 2
Mini-Prep was performed according to standard protocol:
- P803 = pSB1C3_gMK_TK30_SDM-PstI_new clone 1 c = 282,5 ng/µl
- P804 = pSB1C3_gMK_TK30_SDM-PstI_new clone 2 c = 295,6 ng/µl
Test digestion:
| Components | P803 + P804 / µl |
| DNA | 2,5 |
| Buffer 4 | 1 |
| BSA (10x) | 1 |
| PstI | 0,3 |
| H2O | 5,2 |
| Total volume | 10 |
Gel:
0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes
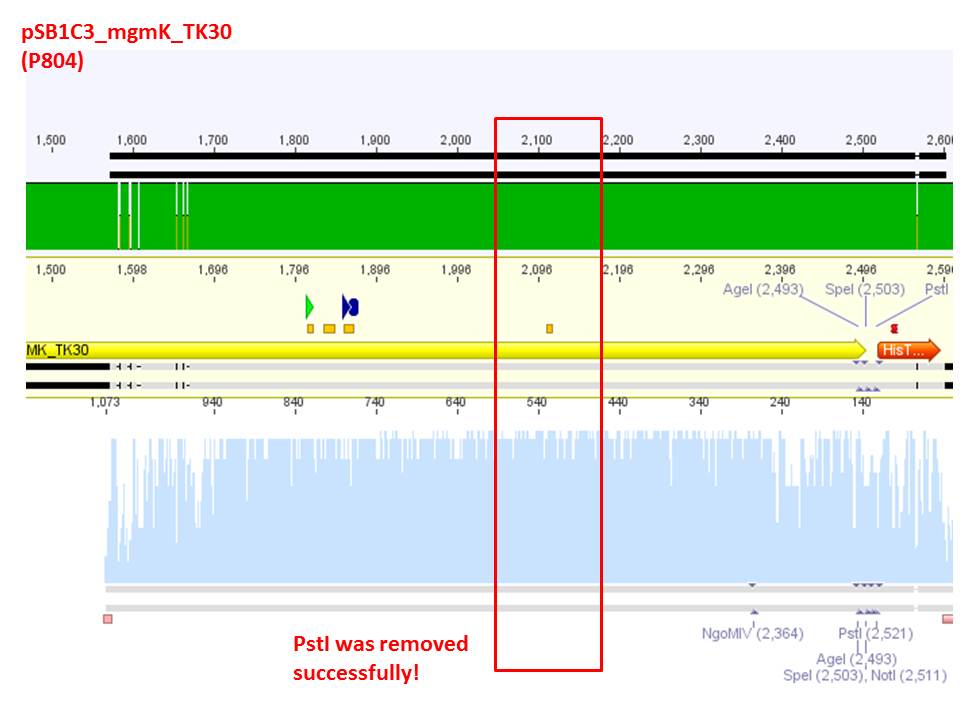
Comment: Vector was cut using PstI in order to verify successful deletion of the PstI restriction site in TK30. As it can be seen above, both vectors are linearized, no fragment was cut out. In case of failed SDM, one additional band at roughly 450 bp was expected. Since this band does not show up, successful deletion is confirmed.
Cloning of lITR_phTERT_beta-globin_mGMK_TK30 into pSB1C3_hgH_rITR
Investigator: Stefan
Comment: In order to test our GOI with another promotor, lITR_phTERT_beta-globin_mGMK_TK30 needs to be cloned upstream of hgH_rITR.
Vector name:
pSB1C3_hgH_rITR (P728)
Insert name:
pSB1C3_lITR_phTERT_beta-globin_mGMK_TK30 (P670)
Digestion:
| components | volume P670 /µl | volume P728 /µl |
| DNA | 10 | 7,7 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme EcoI | 1 | 1 |
| Enzyme XbaI | - | 1 |
| Enzyme SpeI | 1 | - |
| H2O | 4 | 6,3 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
Gel extraction:
Was performed according to protocol.
T4 Ligation:
| ligation name | 728 + 670 |
| volume of vector | 2,35 |
| volume of insert | 5,65 |
| T4 ligase buffer (10x) | 1 |
| T4 ligase | 1 |
Transformation:
Was performed according to standard protocol using BL21 cells.
Cloning of pSB1C3_lITR_phTERT_beta-globin_CFP into pSB1C3_hgH_rITR
Investigator: Anna
Comment:
Insert name:
- pSB1C3_lITR_phTERT_beta-globin_CFP (P791)
Vector name:
- pSB1C3_hgH_rITR (P728)
New vector name:
- pSB1C3_lITR_phTERT_beta-globin_CFP_hgH_rITR
| components | volume for inserts (P666 + P728) /µl | volume of vectors P437 + P715 + P719 /µl |
| DNA | 7,7 | 6,3 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme EcoRI | 1 | 0,7 |
| Enzyme XbaI | 1 | - |
| Enzyme SpeI | - | 0,7 |
| Enzyme BmpI | - | 0,7 |
| H2O | 6,3 | 7,6 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
Gel extraction:
Was performed according to protocol.
| Sample | Vector | Insert |
| DNA concentration [ng/µl] | 19,6 | 14,6 |
| Fragment size [bp] | 2600 | 1800 |
T4 Ligation:
Volume(pSB1C3_hGH_rITR)= 6 µl
Volume (lITR_phTERT_beta-globin_CFP_hgH_rITR) = 2 µl
Transformation:
Was performed according to standard protocol using BL21 cells.
New ligation approach of Rep52
Investigator: Kira
Since ligation with Quickligase (because of one blunt end) revealed no colonies on the plate, I decided to perform ligation with T4 ligase.
Transformation according to the standard protocol with BL21 cells
146. labday 11.10.2010
Colony PCR of Cloning VP2 Fusion and Super constructs into pSB1C3
Investigator: Achim, Hanna
Comment: Because yesterday's cloning didn't deliver a good separartion of the expected gel bands. Nevertheless ligation and trafo was performed. In order to immediately find out, whether we received successful results a colony PCR will be performed.
Two clones were picked from each plat. In addition to that a positive (pAAV_RC) and a negative control (pSB1C3_lITR) was prepared.
Used primer: 4200 rev and Cap3500 for. Expected fragment size: 885 bp.
PCR was performed following the standard protocol.
All samples match the positive control!
To do: Mini-Prep and sequencing.
Preparation of SDS-PAGE gel (10%)
Investigator: Hanna
Comment: In order to perform a Western Blot of different virus capsids (with and without capsid-motifs), 2 10% SDS polyacrylamid gels were prepared.
Resolving gel: 15 mL
- H2O: 5.9 mL
- Acryl-bisacrylamide mix (30%): 5 mL
- Tris (1.5 M, pH 8.8): 3.8 mL
- SDS (10%): 0.15 mL
- Ammonium persulfate (10%): 0.15 mL
- TEMED: 0.006 mL
Stacking gel (5%): 5 mL
- H2O: 3.4 mL
- Acryl-bisacrylamide mix (30%): 0.83 mL
- Tris (1.5 M, pH 6.8): 0.63 mL
- SDS (10%): 0.05 mL
- Ammonium persulfate (10%): 0.05 mL
- TEMED: 0.005 mL
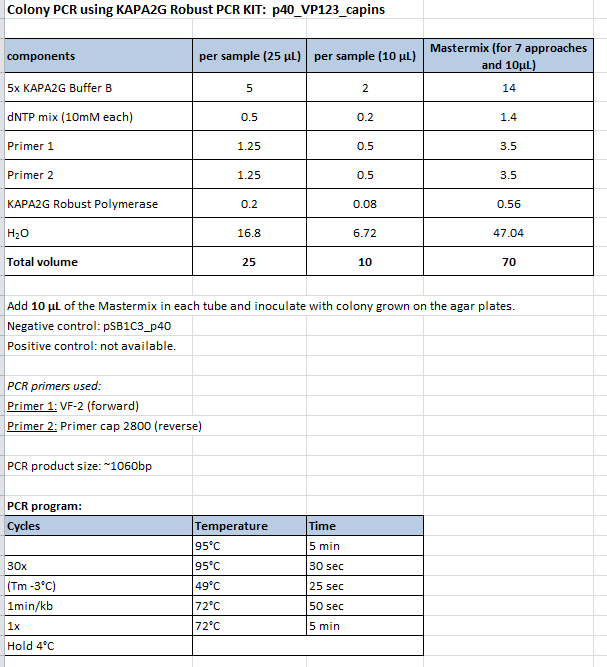
Colony PCR of p40_VP123_capins
Investigator: Bea
Comment: Since the first attempt did not work,and no cells grew on the plate and the same ligation was transformed again into BL-21 and a lot of clones grew on the plate, I decided to perform a colony PCR in order to check several colonies and to inoculate at the same day for a Midi-Prep.
Protocol:
- Primer used: O162
- Primer used: O38
The PCR products were loaded on a 1% agarose gel. The results can be seen above in the gel picture:
Result: We can see two things: The cloning of p40 to VP123 worked quiet well AND the Robust PCR Kit which was used for the first time worked as well.
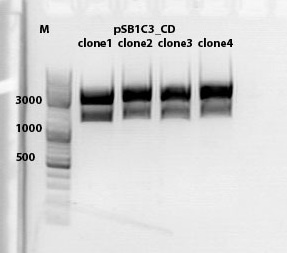
Cloning of lITR_CMV_betaglobin and lITR_phTERT_betaglobin into pSB1C3_CD
Investigator: Stefan
Comment: To produce another GOI for testing in cell culture, the cytosine deaminase needs to be assembled with lITR_promotor_betaglobin. In the next step hgH_rITR needs to be added.
Vector name:
pSB1C3_CD clone 1
pSB1C3_CD clone 2
Insert name:
pSB1C3_lITR_CMV_beta-globin (P729)
pSB1C3_lITR_phTERT_beta-globin (P730)
Digestion:
| components | volume CD clone 1 + 2 /µl | volume P729 /µl | volume P730 /µl |
| DNA | 6 | 14 | 6 |
| BSA (10x) | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 |
| Enzyme EcoI | 1 | 1 | 1 |
| Enzyme XbaI | 1 | - | - |
| Enzyme SpeI | - | 1 | 1 |
| H2O | 8 | - | 8 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 | 20 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
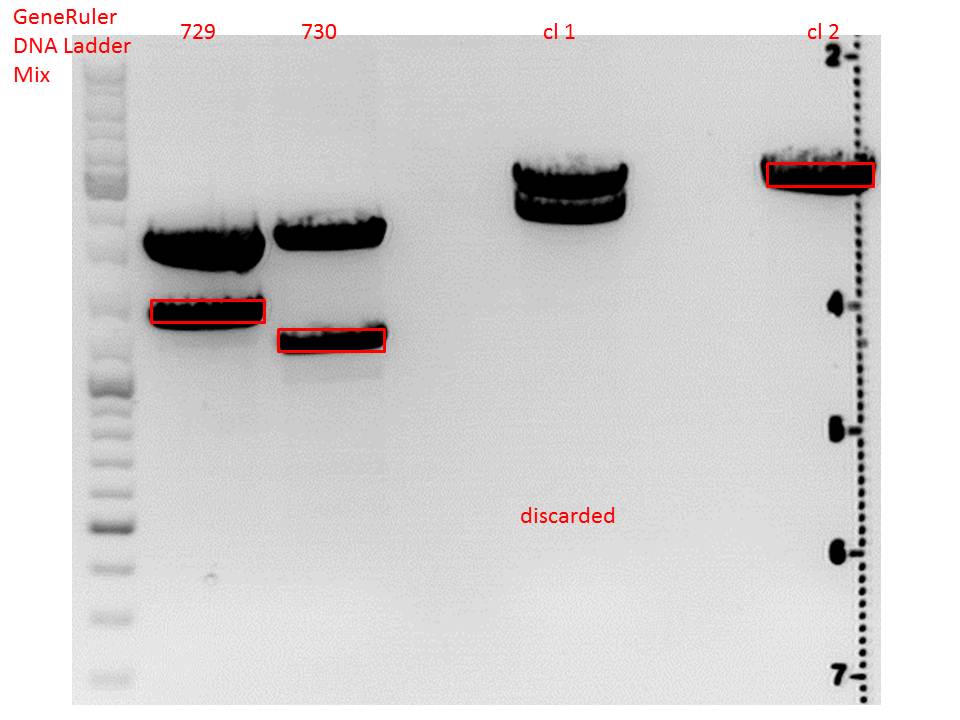
CD clone 1 yielded to bands around 2500 bp to 3000 bp. Since the vector was cut only using EcoRI and SpeI, it was expected to be linearized, not to be cut into two fragments this size. Therefore, this sample was discarded and cloning was continued using CD clone 2.
Gel extraction:
Was performed according to protocol.
T4 Ligation:
| ligation name | 729 + CD cl2 | 730 + CD cl2 |
| volume of vector | 3,67 | 2,82 |
| volume of insert | 4,33 | 5,18 |
| T4 ligase buffer (10x) | 1 | 1 |
| T4 ligase | 1 | 1 |
Transformation:
Was performed according to standard protocol using BL21 cells.
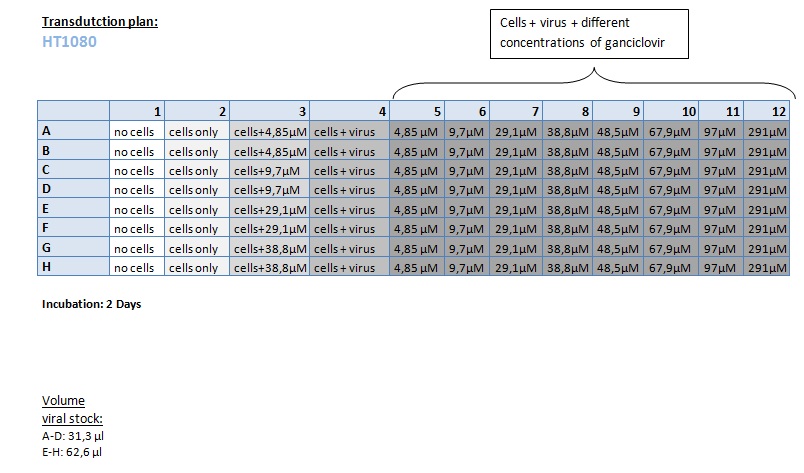
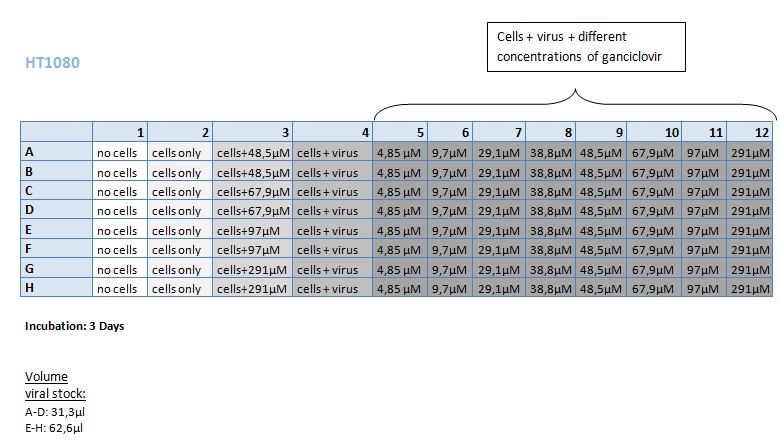
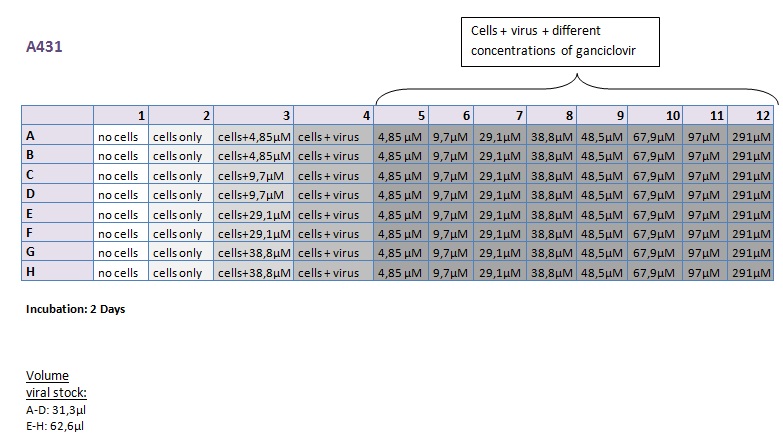
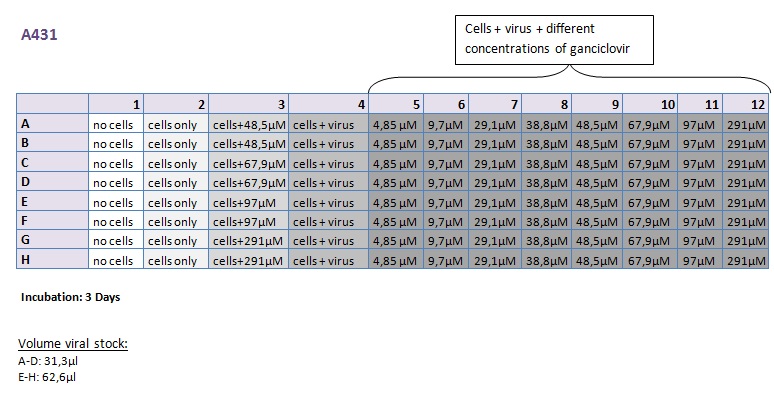
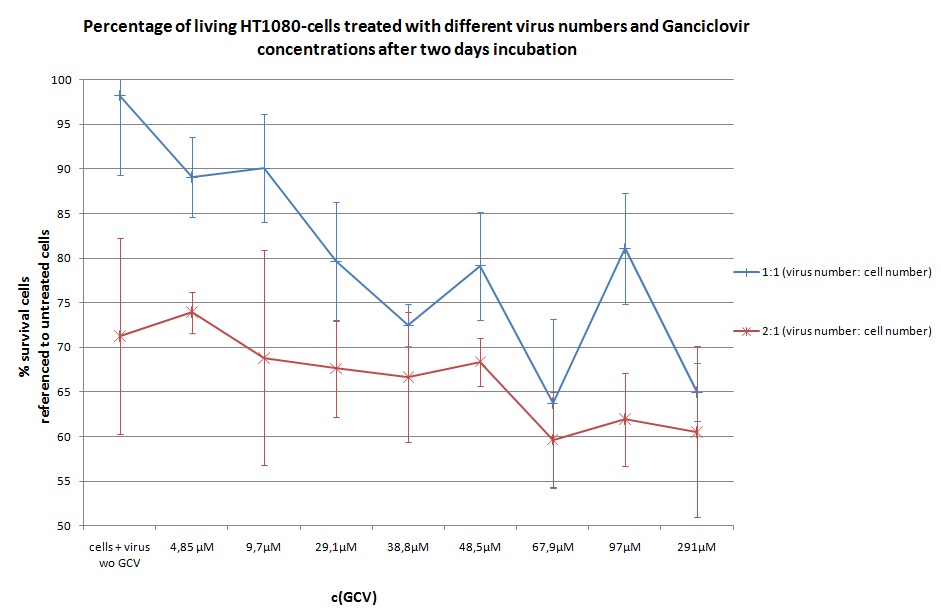
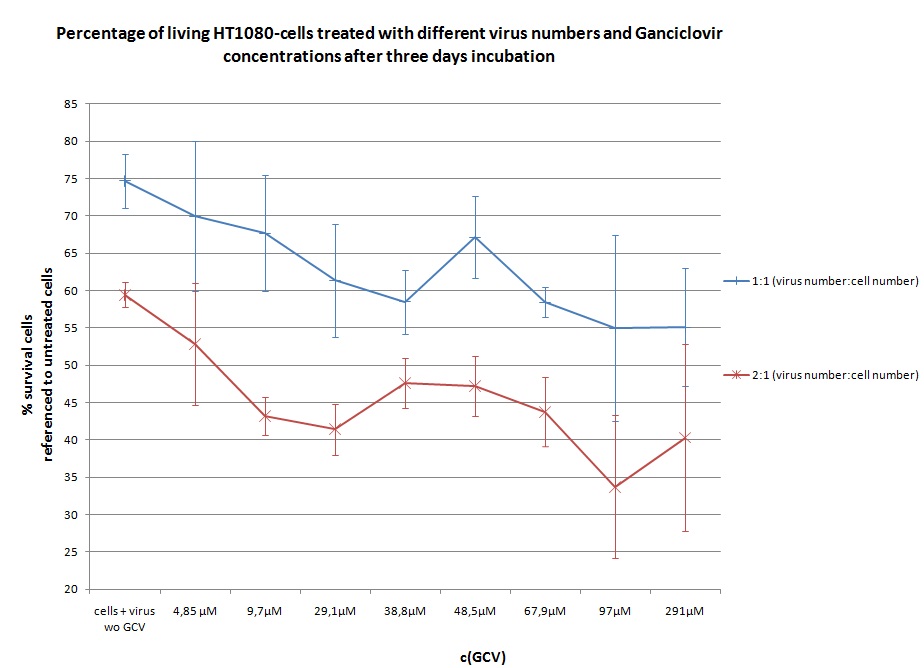
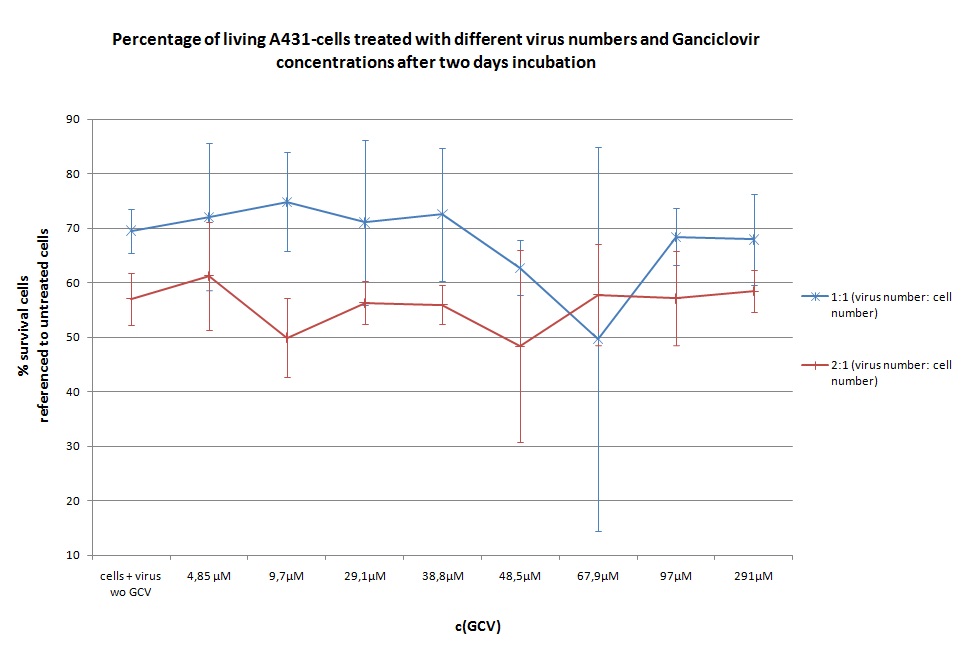
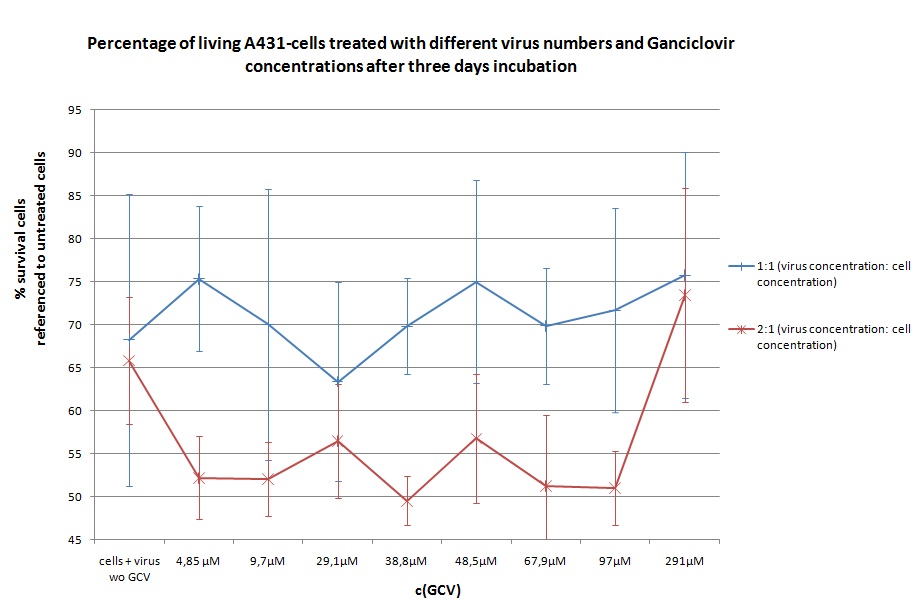
Seeding HT1080 and A431 for testing different concentrations of ganciclovir by MTT-Assay
Investigator: Kerstin, Anissa
- Seeding 4x 96-well plates: 2x HT1080 and 2x A431
FACS-Analysis
Investigator: Kerstin
...
Preparation of the ELISA
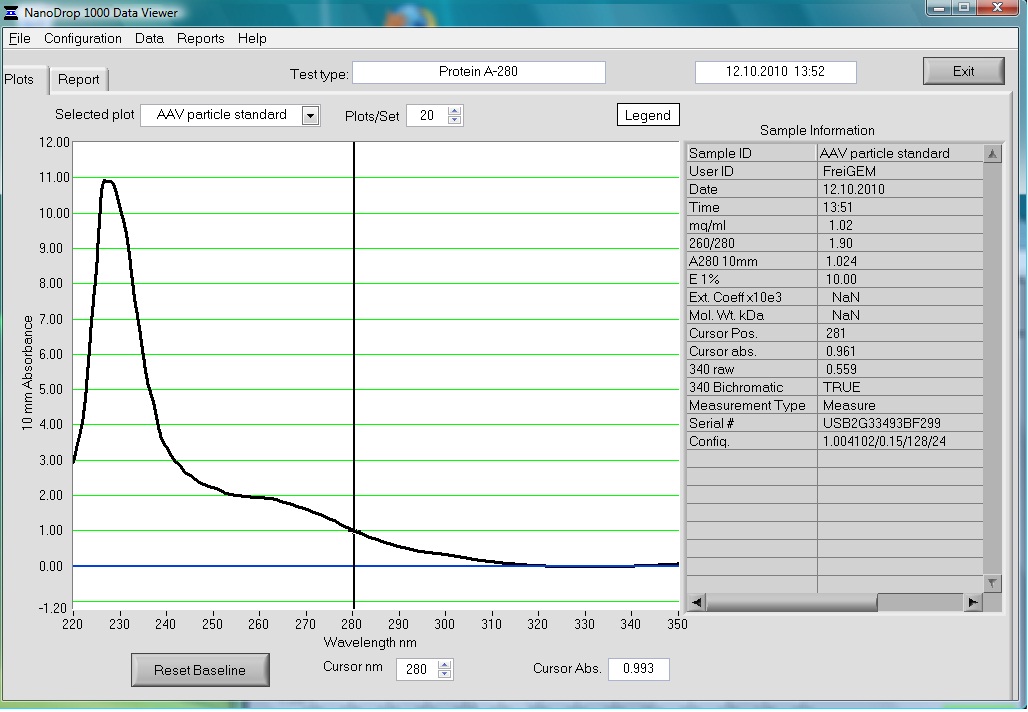
Investigator: Volker
The AAV particle standard that contains 3.6x10^9 viral particles was dissolved in 500µl as described in the protocol of the Progen AAV Titration ELISA and a absorption spectrum was measured.
These purified and concentrated viral particles could be used for biophysical measurements, there for the possibility to detect the viral particles by absorption was interesting for us.
The Spectrum measured in the NanoDrop is the following:
Trafo evaluation of Rep52
Investigator:Kira
T4 ligation seems to worked out. The plate contained many colonies.
147. labday 12.10.2010
Sequencing analysis: pSB1C3_mGMK_TK30 and pSB1C3_CD
Investigator: Stefan
Test digestion of pSB1C3_CD and pSB1C3_CD_SDM-PstI_new
Investigator: Stefan
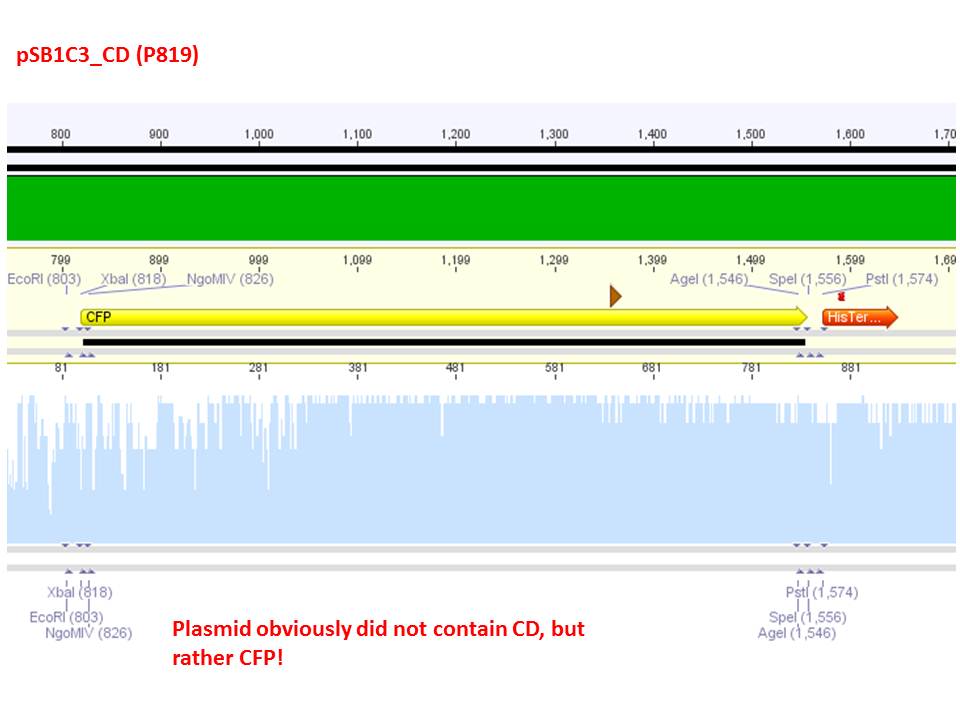
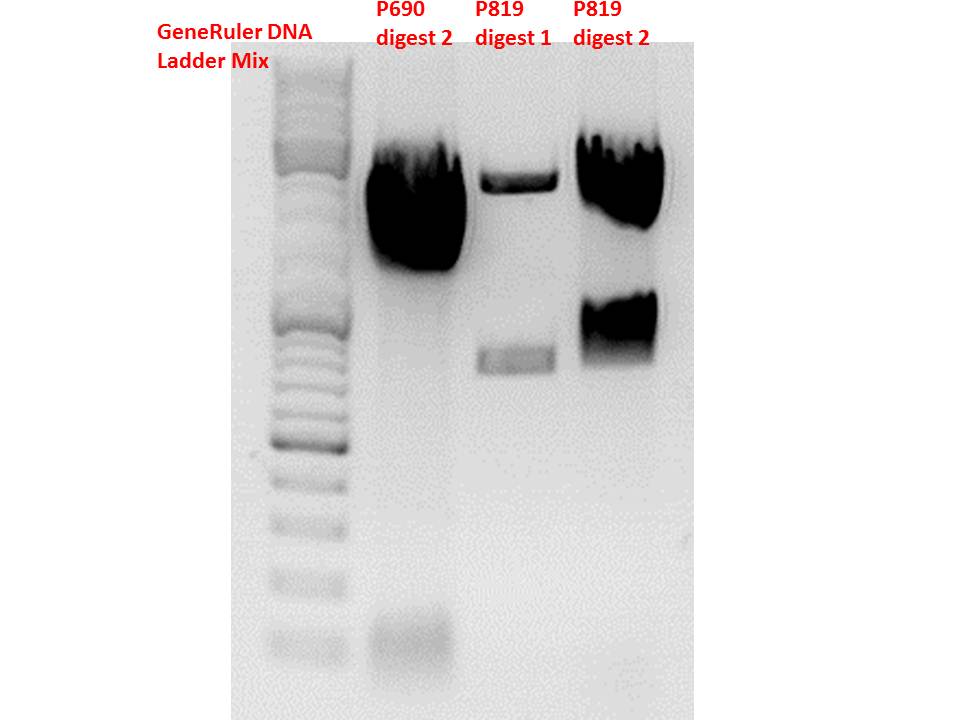
Comment: Sequencing results revealed that there is CFP rather than CD in P819. In order to verify this and exclude the possibility that the wrong plasmid was sent for sequencing test digestions were performed. Additionally the original vector at which the SDM to remove the PstI was performed was digested as well to verify its insert.
| Components | digest 1 / µl | digest 2 / µl |
| DNA | 1 | 7 |
| Buffer 4 | 1 | 1 |
| BSA (10x) | 1 | 1 |
| Enzyme I | SpeI 0,3 | PstI 0,3 |
| Enzyme II | XbaI 0,3 | XbaI 0,3 |
| H2O | 6,4 | 0,4 |
| Total volume | 10 | 10 |
Comment: Using these digestion approaches, it was expected for CFP to deliver always bands at ~750 bp. For the CD it was expected to show bands at 90 bp, 1200 bp and 2000 bp if the CD still contains the PstI restictiton site, 1300 bp and 2000 bp if it was removed.
Gel:
0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes
Comment: As it can be seen, P819 does contain CFP, therefore a mistakenly sequenced plasmid can be excluded. Also it can be seen that P690 contains the CD. Therefore an additonal SDM of P690 to remove the PstI restriction site is necessary.
qPCR of virus stock: P816/ TKGMk-Standard; R/C: P468 from 5.10. (VLP harvested by Adrian)
Investigator: Achim
Protocol for quantitative real-time PCR of virus particles
Date: 12.10.
Achim
Virus Stock: A.F.; P816; TKGMK-Standard; R/C: P468; 5.10; consists of supernatant from pelleted cell fragments.
Following the Protocol used by (Rohr et al., 2002).
I digested 5 µl virus dilution with 7,5 µl DNAse I (7,5 units) and 25 µl 50 mM MgCl2 (end concentration 25 mM) in a final volume of 50 µl at 37°C for 30 min.
è DNAse should be heat inactivated at 65°C for 10 min. I forgot that step. The enzyme should however be inactivated in the initial denaturation step.
I prepared the following PCR reactions:
|
Sample |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|
9 |
10 |
|
|
1.1 |
1.2 |
2.1 |
2.2 |
S.1 |
S.2 |
NP1 |
NP2 |
MM for samples 1 -8 |
NH1 |
NH2 |
|
PCRmix |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
112.5 |
12.5 |
12.5 |
|
Primer for |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
9 |
- |
- |
|
Primer rev |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
9 |
- |
- |
|
Template |
2 |
2 |
2 |
2 |
5 |
5 |
- |
- |
- |
- |
- |
|
H2O |
8.5 |
8.5 |
8.5 |
8.5 |
5.5 |
5.5 |
10.5 |
10.5 |
- |
12.5 |
12.5 |
|
Total |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
|
25 |
25 |
|
Step |
Time |
Temp |
|
Initial denaturation |
7’ |
95°C |
|
|
|
|
|
Denaturation |
10’’ |
95°C |
|
Annealing/Extension |
30’’ |
60°C |
|
Number of Cycles |
45 |
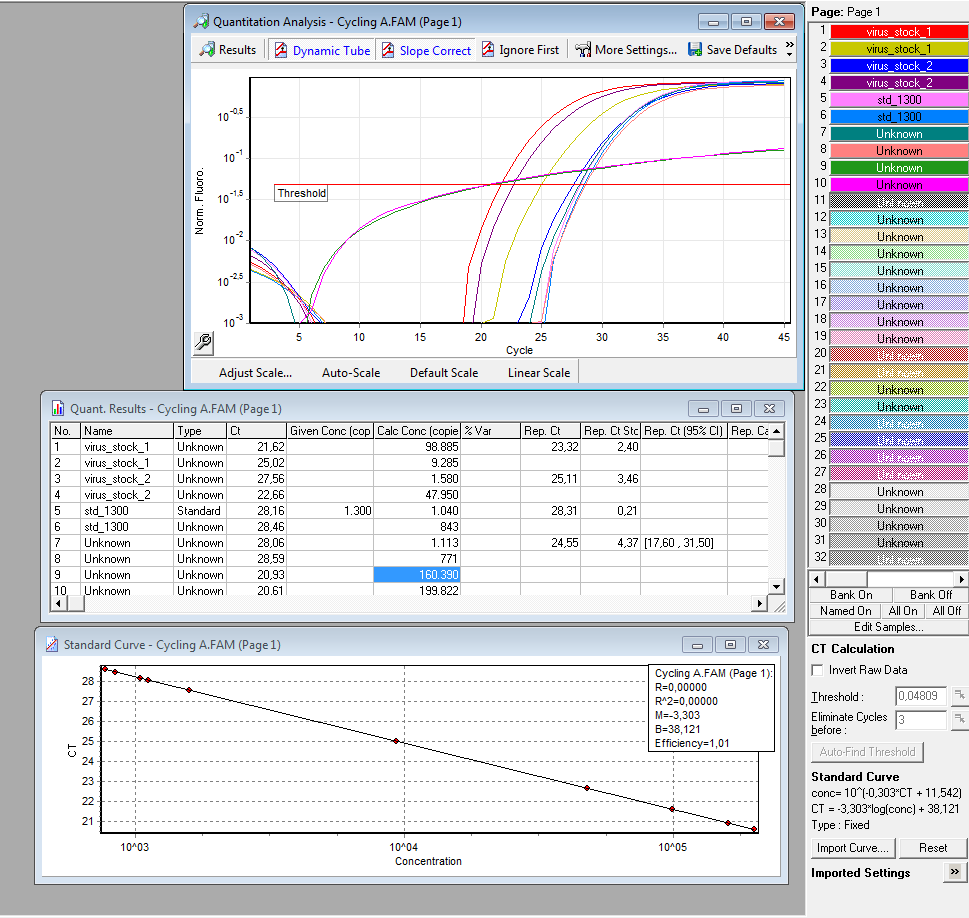
|
Results:
è Both negative controls containing primers were contaminated with DNA.
è Negative controls containing only H2O were negative.
è Values for plasmids/reaction vary; from 1 – 100 for the same virus stock
è Dilution of the virus solution: 1:10; 2µl PCR sample: 3.2 * 10^5 virus particles/ml
è Because of the contaminated negative controls and the high deviation, I will repeat the experiment tomorrow to verify the results.
References:
Rohr, U., Wulf, M., Stahn, S., Steidl, U., Haas, R., Kronenwett, R., et al. (2002). Fast and reliable titration of recombinant adeno-associated virus type-2 using quantitative real-time PCR. Journal of virological methods, 106(1), 81-8. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12367732.
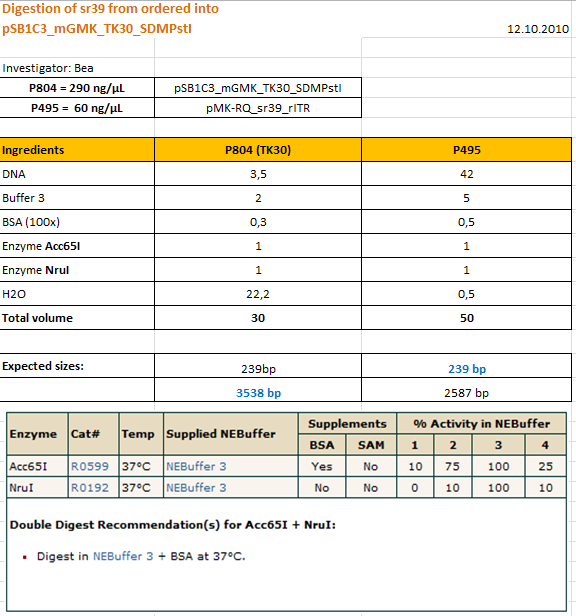
Cloning sr39 into mGMK_TK30
Investigator: Bea
Comment: We do not only want to submit the tk30, a mutant version of the thymidine kinase, but as well another mutant protein which is named sr39. SR39 seems to have a better activity than the tk30 without being fused to the guanylate kinase (mgmk), but the sr39 also works quite well as a fusion protein. We ordered one part of the sr39 and we are now subcloning it into the mgmnk_tk30 construct in order to obtain mgmk_sr39.
Protocol:
The digested fragments were loaded on a 1.2% agarose gel. The results can be seen above in the gel picture: I cut out the small visible band in the right lane, and the upper intensive band of the left lane.
Result:
After gel extraction has been performed, I ligated the two fragments and transformed BL-21 cells, plated them on cm agar plates and incubated them over night at 37°C.
Concentration of virus stock for Western Blot analysis
Investigator: Hanna
For the first Western Blot try, we investigate whether is is enough to concentrate the virus stock (from cell supernatant) and then to simply load it onto a SDS-PAGE gel and perfom immuno-staining.
For this purpose 8 mL virus stock (pHelper, pAAV_RC, GOI=YFP) were concentrated via amicon filtration.
- First, amicon filter was washed with 4 mL PBS.
- Then 4 mL virus stock was loaded onto the filter and centrifuged at 3000 rpm for 15 minutes.
- Flow-through was discarded and additional virus stock was loaded, centrifuged at 3000 rpm for 25 minutes.
- Flow-through was discarded, remaining solution was re-suspended and additional virus stock was added. Amicon filter was centrifuged at 3000 rpm for 50 minutes.
- Flow-through was discarded. Residual protein solution: 500 µL. Filter was washed with 3 mL PBS, centrifuging 35 minutes two times.
- 2 x 40 µL protein solution (16x) was taken and mixed with Laemmli buffer and stored over night @ 4°C.
To do: SDS-PAGE and Western Blot tomorrow.
Perfomance of another try by harvesting viruses of the cell pellet with RIPA buffer & freeze/ thaw.
Western Blot of unmodified, and modified capsids (N-terminal fusion and VP1 insertion with CFP ref. mVenus) --> size shift should be detectable.
MTT Assay (First Try) Transduction of HT1080 and A431
Investigator: Kerstin, Anissa
Results:
Mini-Prep and test digestion of several constructs
Investigator: Stefan
Glycerol stocks were prepared:
- B651 = pSB1C3_P40_VP123
- B652 = pSB1C3_lITR_phTERT_ß-globin_mGMK_TK30_PstI_SDM_hgH_rITR
- B653 = pSB1C3_lITR_phTERT_ß-globin_mGMK_TK30_PstI_SDM_hgH_rITR
- B654 = pSB1C3_lITR_CMV_ß-globin_mGMK_TK30_hgH_rITR
- B655 = pSB1C3_lITR_CMV_ß-globin_mGMK_TK30_hgH_rITR
- B656 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO
- B657 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO
- B658 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO
- B659 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO
- B660 = pSB1C3_lITR_phTERT_ß-globin_CFP_hgH-rITR
- B661 = pSB1C3_lITR_phTERT_ß-globin_CFP_hgH-rITR
Mini-Prep was performed according to standard protocol:
- P821 = pSB1C3_P40_VP123 c = 327,4 ng/µl
- P822 = pSB1C3_lITR_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR c = 342,9 ng/µl
- P823 = pSB1C3_lITR_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR c = 324,0 ng/µl
- P824 = pSB1C3_lITR_CMV_ß-globin_mGMK_TK30_hgH_rITR c = 81,6 ng/µl
- P825 = pSB1C3_lITR_CMV_ß-globin_mGMK_TK30_hgH_rITR c = 148,4 ng/µl
- P826 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO c = 105,8 ng/µl
- P827 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO c = 121,1 ng/µl
- P828 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO c = 188,4 ng/µl
- P829 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO c = 140,4 ng/µl
- P830 = pSB1C3_lITR_phTERT_ß-globin_CFP_hgH-rITR c = 197,2 ng/µl
- P831 = pSB1C3_lITR_phTERT_ß-globin_CFP_hgH-rITR c = 207,3 ng/µl
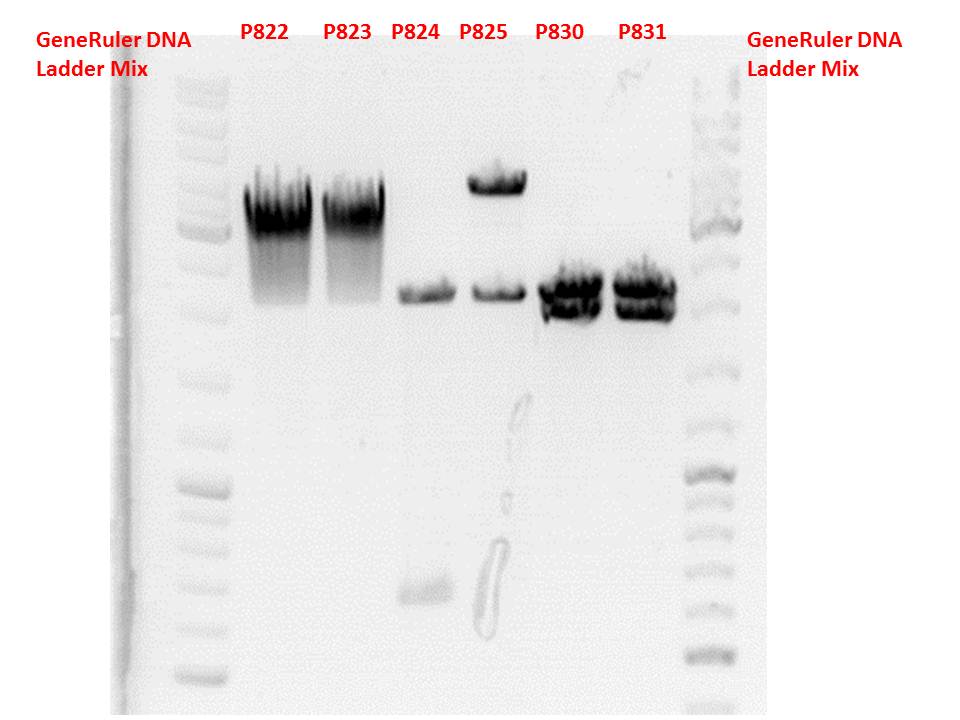
Test digestion:
| Components | P822 + P823 / µl | P824 + P825 / µl | P830 + P831 / µl |
| DNA | 1,5 | 1,5 | 2 |
| Buffer 4 | 1 | 1 | 1 |
| BSA (10x) | 1 | 1 | 1 |
| PstI | 0,3 | - | - |
| XbaI | 0,3 | 0,3 | 0,3 |
| SpeI | - | 0,3 | 0,3 |
| H2O | 5,9 | 5,9 | 5,4 |
| Total volume | 10 | 10 | 10 |
Gel:
0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes
Comment: P822 and P823 smear a lot, therefore evaluation is difficult. Another test digestion would be reasonable.
P824 contains hgH_rITR, therefore it will be discarded. P825 looks well and was sent for sequencing.
P830 and P831 look well and will be sent for sequencing tomorrow.
Repetiton: Site-directed mutagenesis in pSB1C3_CD construct
Investigator: Stefan
Comment: Sequencing revealed that there is no CD in our pSB1C3_CD but rather CFP. So, anonther approach of the SDM needs to be performed.
Protocol using the QuickChange Lightning Kit from Stratagene:
Ingredients:
| Ingredients | Volume / µl |
| 10x reaction buffer | 2,5 |
| dNTP | 0,5 |
| forward primer: O191 | 0,56 |
| reverse primer: O192 | 0,57 |
| DNA Template | 0,5 (1:10 dilution) |
| QuikSolution Reagent | 0,75 |
| QuikChangeLightning Enzyme | 0,5 |
| H2O | 14,35 |
| Total volume | 25 |
The following PCR program was used:
- 95 °C 120 s
- 95 °C 20 s
- 60 °C 60 s
- 68 °C 105 s
- Goto step 2 repeat 17 times
- 68 °C 300 s
Digestion using DpnI:
To digest methylated and hemi-methylated DNA, 1 µl of DpnI was added and the mixture was incubated for 10 minutes at 37 °C.
Transformation:
Trafo was performed according to protocol, but using XL1b cells instead of XL-10 Gold cells.
148. labday 13.10.2010
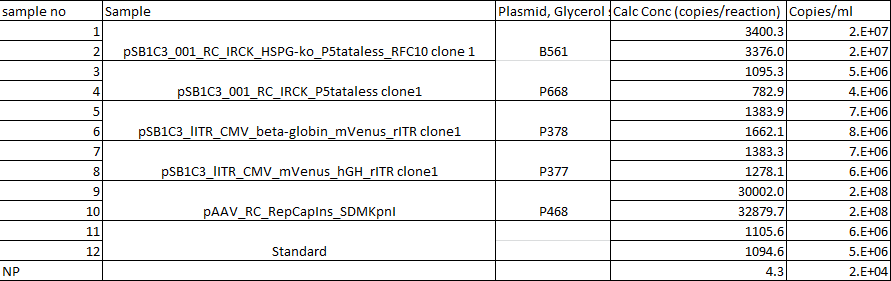
qPCRs of virus stocks
Investigator: Achim
|
Sample: |
Plasmid, Glycerol stock: |
Sample No. |
|
pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10 clone 1 |
B561 |
1 |
|
pSB1C3_001_RC_IRCK_P5tataless clone1 |
P668 |
2 |
|
pSB1C3_lITR_CMV_beta-globin_mVenus_rITR clone1 |
P378 |
3 |
|
pSB1C3_lITR_CMV_mVenus_hGH_rITR clone1 |
P377 |
4 |
|
pAAV_RC_RepCapIns_SDMKpnI |
P468 |
5 |
Digestion: 37°C, 30 min
|
|
1 |
2 |
3 |
4 |
5 |
MM x6 |
|
Virus solution |
5 |
5 |
5 |
5 |
5 |
|
|
DNAseI (7,5 U) |
7,5 |
7,5 |
7,5 |
7,5 |
7,5 |
45 |
|
MgCl2 50 mM |
5 |
5 |
5 |
5 |
5 |
30 |
|
H2O |
32.5 |
32.5 |
32.5 |
32.5 |
32.5 |
195 |
|
Total |
50 |
50 |
50 |
50 |
50 |
300 |
PCR reaction:
|
Sample |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|
15 |
16 |
|
|
1.1 |
1.2 |
2.1 |
2.2 |
3.1 |
3.2 |
4.1 |
4.2 |
5.1 |
5.2 |
S.1 |
S.2 |
NP1 |
NP2 |
MM for samples 1 -14 (x15) |
NH1 |
NH2 |
|
PCRmix |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
187,5 |
12.5 |
12.5 |
|
Primer for 1:10 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
15 |
- |
- |
|
Primer rev 1:10 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
15 |
- |
- |
|
Template |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
5 |
5 |
- |
- |
- |
- |
- |
|
H2O |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
5.5 |
5.5 |
10.5 |
10.5 |
- |
12.5 |
12.5 |
|
Total |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
|
25 |
25 |
Results:
SDS Page of unmodified AAV2 capsids
Investigator: Hanna
Comment: Yesterday a 8x concentration of AAV2 supernatant-stock was performed. 10 µL Laemmli-buffer was added to 40 µL AAV stock (8x) and stored over night at 4°C.
Today a SDS-PAGE of these samples will be performed.
Samples were incubated for 10 minutes at 95°C for denaturation.
Loading plan:
| 5 µL PageRuler Prestained Protein Ladder | 10 µL Sample | 15 µL Sample | 25 µL Sample |
After running front reached the end of the gel, PAGE was stopped.
Western Blot:
Three layers of transfer buffer soaked whatman paper were placed onto the blotter. A polyvinylidene difluoride membrane was activated by methanol (5 seconds) and then incubated for 5 minutes in transfer buffer. Membrane was then placed onto the whatman paper, gel was carefully layed on top. Air bubbles were removed, ensuring elictricity flow. After coverage by 3 further layers of whatman, blotting was performed at 200 mA for 1 hour.
Blocking:
In order to minimize nonspecific antibody interactions, membrane was incubated in blocking buffer (TBS (pH 7.5), 5% milk powder) for one hour, shaking.
Primary antibody incubation:
A 1:10 dilution of the B1 antibody in TBS + 1% milk was prepared. Membrane was incubated in 50 mL falcon over night at 4°C, rotating.
Midi-Prep
Investigator: Chris W.
Midi-Prep of:
pSB1C3_CMV_VP123_453_Z34C =P832 =B656
pSB1C3_CMV_VP123_453_RGD_HSPG-KO =P833 =B658
pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10 clone 1 =P834 =B561
pSB1C3_001_RC_IRCK_P5tataless clone 1 =P835 =B516
pSB1C3_lITR_CMV_betaglobin_mVenus_hGH_rITR clone1 =P836 =B200
pSB1C3_lITR_CMV_betaglobin_mVenus_hGH_rITR clone1 =P837 =B200
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P832 | P833 | P834 | P835 | P836 | P837 |
| concentration (ng/µl) | 3615,8 | 1504,7 | x | x | x | x |
Comment: P834-837 given to sven
Repetiton: Test digestion of pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_SDM-PstI_hgH_rITR
Investigator: Stefan
Comment: Repetiton of yesterday's test digestion which yielded no clear band at 2000 bp as it was expected.
| Components | P822/P823 / µl |
| DNA | 1,5 |
| Buffer 4 | 1 |
| BSA (10x) | 1 |
| SpeI | 0,3 |
| XbaI | 0,3 |
| H2O | 5,9 |
| Total volume | 10 |
Gel:
0,45g agarose, 50 ml TAE (0,9%), 3 µl GELRED, 90 Volt, running time ~110 minutes
Comment: This time both samples look well. P822 will be sent for sequencing.
Sequencing analysis: pSB1C3_leftITR_pCMV_betaglobin_mGMK_TK30_hgH_rITR (P825)
Investigator: Stefan
For sequencing the following primers were used_
- qPCR primer CMV for (O20)
- GOI primer for (O30)
- seq. primer TK GMK for (O23)
- TK30 SDM Pst for (O191)
Sequencing comfirmed that the pysical sequence fits our theoretical. Next step will be to remove the remaining PstI restriction site within TK30.
Repetition of cloning of Rep into Rep78
Investigator: Kira
Comment: The last 2 approaches to digest Rep78 and to clone failed because Rep78 was almost gone after PCR purification and was not detectable on the gel, even if I used the double amount of DNA. Thus, new over night culture was grown in order to perform the experiment again
Regarding the activity conditions of the enzymes, digestion was performed in 2 steps.
| Components | Rep78 |
| DNA | 3 µl |
| BSA (100x) | 2 µl |
| Buffer no. 3 | 2,0 µl |
| Enzyme SwaI | 1,0 µl |
| H2O | 12 µl |
| Total volume | 20 |
incubation @ 25 C for approx. 2 h
Dephosphorylation was performed because of the blunt cleavage. PCR phosphorylation followed in order to get rid off the buffer because Buffer 3 is not appropriate for further digestion.
| Components | Rep78 |
| DNA | 30 µl |
| BSA (100x) | 0 µl |
| Buffer no. 2 | 4,5 µl |
| Enzyme HindIII | 1,0 µl |
| H2O | 9,5 µl |
| Total volume | 45 |
incubation for 2h at 37C
Ligation with T4 ligase
v(insert)=4,4 ul
v(vector)= 3,6 ul
T4 buffer= 1 ul
T4 ligase= 1 ul
incubation at RT for 45 min
Transformation according to the standard protocol with BL21
Midi-Prep
Investigator: Chris W.
Midi-Prep of:
pSB1C3_CMV_VP123_capins =P862 =B474
pSB1C3_P40_VP123 =P863 =B651
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P862 | P863 |
| concentration (ng/µl) | 3310,5 | 2209,2 |
149. labday 14.10.2010
Mini-Prep and test digestion of several constructs
Investigator: Stefan
Glycerol stocks were prepared:
- B658 = pSB1C3_CD_SDM-PstI_III clone 1
- B659 = pSB1C3_CD_SDM-PstI_III clone 2
- B660 = pSB1C3_mGMK_sr39 clone 1
- B661 = pSB1C3_mGMK_sr39 clone 2
Mini-Prep was performed according to standard protocol:
- P858 = pSB1C3_CD_SDM-PstI_III clone 1 c = 241,7 ng/µl
- P859 = pSB1C3_CD_SDM-PstI_III clone 2 c = 212,1 ng/µl
- P860 = pSB1C3_mGMK_sr39 clone 1 c = 231,8 ng/µl
- P861 = pSB1C3_mGMK_sr39 clone 2 c = 265,2 ng/µl
Test digestion:
| Components | P858 + P859 / µl | P860 + P861 / µl |
| DNA | 7 | 1,5 |
| Buffer 4 | 1 | 1 |
| BSA (10x) | 1 | 1 |
| Enzyme I | PstI 0,3 | BstXI 0,3 |
| Enzyme II | EcoRI 0,3 | XbaI 0,3 |
| Enzyme III | - | SpeI 0,3 |
| H2O | 0,4 | 5 |
| Total volume | 10 | 10 |
Gel:
0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes
Comment: .
Continuation of Western Blot of unmodified AAV2 capsids
Investigator: Hanna
Comment: Yesterday, SDS PAGE, Western Blot and primary B1 mouse antibody incubation was performed.
Today, secondary antibody incubation and ECL analysis will be done.
- Membrane was washed 3 x 7 minutes in TBST (0.2% Tween).
- A 1:5000 dilution of secondary goat anti-mouse HRP (sc2005) coupled antibody in TBS was prepared.
- Membrane was incubated in secondary antiobody for 1 hour.
- Membrane was washed 3 x 10 minutes.
- Pierce ECL Western Blotting Substrate was prepared: 350 µL Luminol Enhancer Solution + 350 µL Peroxidase Solution
- Analysis of the membrane revealed that the samples didn't run well. Much unspecific antibody binding was detectable.
Because of these dissatisfying results SDS PAGE will be repeated. Instead of performing Western Blot, gel wioll be stained by Coomassie in order to optimize SDS-PAGE conditions.
Cloning hGH_rITR into (form P728) into pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_SDM-PstI (P822)
Investigator Patrick
Digestions, 1 h 40 minutes, 37 °C:
- P728: 1 µl XbaI, 1 µl PstI, 2 µl BSA, 2 µl Buffer 4, 6 µl DNA, 8 µl H2O
- P822: 1 µl SpeI, 1 µl PstI, 2 µl BSA, 2 µl Buffer 4, 6 µl DNA, 8 µl H2O
Expected results for the 1% agarose gel:
- P728: about 2100 and 650 bp
- P822: about about 4900 and 10 bp (wont be visible on the gel)
As usual the PSB1C3 bands indicate a higher bp amount than they actually got.
The gelextraction ...
- P728: 9 ng/µl
- P822: 67,5 ng/µl
... and following ligation (2 µl Insert, 6 µl vector, 1 µl T4 DNA Ligase, 1 µl T4 DNA Ligase Buffer, 40 minutes, RT) were performed according to the standard protocol. After the transformation (with BL21) the cells were plated and put into the 37°C room.
Cloning of the ViralBricks 453-empty and 587 empty
These ViralBricks were not ordered as hybridized oligos, but can be cloned from the Cap-Gene-Synthesis into the ViralBrick MCS.
| components | volume of p320 (ViralBrick-BLA) /µl | volume of p320 (ViralBrick-BLA) /µl | volume of p668 (pSB1C3_RepCap_IRCK) /µl | volume of p668 (pSB1C3_RepCap_IRCK) /µl |
| DNA | 2µl | 2µl | 7µl | 7µl |
| BSA (100x) | 2.5µl | 2.5µl | 2.5µl | 2.5µl |
| Buffer 4 (10x) | 2.5µl | 2.5µl | 2.5µl | 2.5µl |
| SspI | 1µl | 0 | 1µl | 0 |
| SalI | 1µl | 0 | 1µl | 0 |
| BamHI | 0 | 1µl | 0 | 1µl |
| PvuII | 0 | 1µl | 0 | 1µl |
| H2O | 16µl | 16µl | 11µl | 11µl |
| Total volume | 25µl | 25µl | 25µl | 25µl |
| Sample Marker | Sample ViralBrick_BLA 453 cut | Sample ViralBrick_BLA 587 cut | Sample pSB1C3_001_RepCap_IRCK 453 cut | Sample pSB1C3_001_RepCap_IRCK 587 cut | |
|---|---|---|---|---|---|
| Lane | 7µl | 30µl | 30µl | 30µl | 30µl |
SDS PAGE of viral capsids
Investigator: Hanna
Comment: In order to optimize SDS PAGE with the viral capsid proteins, I didn't perform Western Blot, but Coomassie staining.
1.: Preparation of 10% polyacrylamide gel:
Following standard protocol.
2.: Preparation of virus samples:
Two different virus stocks with unmodified capsid were taken. 2 samples were prepared of each: One sample was untreated, the other one was centrifuged in order to sediment cell fragments.
10 µL Laemmli buffer (5x) was added to 40 µL virus sample and incubated at 95°C for 5 minutes.
3.: SDS PAGE
Loading plan:
| 10 µL PageRuler Prestained Protein Ladder | 10 µL Sample 1 (not centrifuged) | 20 µL Sample 1 (not centrifuged) | 10 µL Sample 1 (centrifuged) | 20 µL Sample 1 (centrifuged) | 10 µL Sample 2 (not centrifuged) | 20 µL Sample 2 (not centrifuged) | 10 µL Sample 2 (centrifuged) | 20 µL Sample 2 (centrifuged) |
Gel was run 1.5 hours at 10 mA.
4.: Coomassie staining
Gel was put into coomassie bath, heated in microwave and incubated for 1 hour, shaking.
In order to remove background coomassie staining, gel was incubated in acetic acid (20%) for 2 hours, shaking.
Results revealed that samples ran well (vertically :) ) - 20 µL seems to be too much. Interestingly centrifuging the virus stocks in order to remove cell debris didn't lead to better results.
To do: Perform Western Blot again - with 10 µL Virus stock, 0.5% instead of just 0.2% TBS-milk and longer washing steps.
Sequence analysis of super constructs
Investigator: Hanna
Comment: We assembled targeting super constructs which specifically bind the EGF receptor with VP2 N-terminal fused ZEGFR:1907 Affibody and can either be purified via a loop inserted His-Tag or visualized via Streptavidin coupled fluorescent proteins.
Further on these loop motifs were vloned to the "mVenus VP1 insertion".
Sequencing results showed that insertion of these constructs into pSB1C3_CMV worked!
Next steps: Put sample sinto BioBrick Box.
Midi-Prep + Transfection.
Transfection with pCMV, p40, pAAV_RC
Investigator: Kira
Comment: Transfection was performed in order to evaluate the promoter activities.
Yesterday, the cells were seeded in the 6 well plate by Sven. Transformation was performed according to the manufacturer protocol (Qiagen).
Trafo evaluation of Rep78
Investigator: Kira
Transformation was successful. 2 clones will be picked for mini prep tomorrow.
Picking clones
Investigator: Kira
Rep78 and Rep52 were picked (2 clones from each plate), in order to continue with mini prep and PCR amplification.
Cloning of CD into pSB1C3_hGH_rITR
Investigator: Stefan
.
Vector name:
pSB1C3_hGH_rITR (P728)
Insert name:
pSB1C3_CD_SDM-PstI_III clone 1 (P858)
pSB1C3_CD_SDM-PstI_III clone 2 (P859)
Digestion:
| components | volume P858 + P859 /µl | volume P728 /µl |
| DNA | 6 | 8 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme EcoI | 1 | 1 |
| Enzyme XbaI | - | 1 |
| Enzyme SpeI | 1 | - |
| H2O | 8 | 6 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
Gel extraction:
Was performed according to protocol.
T4 Ligation:
| ligation name | 728 + 858 | 728 + 859 |
| volume of vector | 3,14 | 3,14 |
| volume of insert | 4,86 | 4,86 |
| T4 ligase buffer (10x) | 1 | 1 |
| T4 ligase | 1 | 1 |
Transformation:
Was performed according to standard protocol using BL21 cells.
 "
"