Team:Freiburg Bioware/NoteBook/Labjournal/October
From 2010.igem.org
(→Impressions of transfection of AAV293 with pCerulean_VP1up_NLS_mVenus_VP2/3_insCap) |
(→Biobrick assembly pSB1C3_lITR_hTERT_beta-globin_CD) |
||
| Line 72: | Line 72: | ||
'''Ligation'''<br /> | '''Ligation'''<br /> | ||
| - | DNA-mix: 8 ul (vector | + | DNA-mix: 8 ul (vector 4,6ul)+(insert 3,4 ul) <br /> |
T4 ligase: 1 ul <br /> | T4 ligase: 1 ul <br /> | ||
T4 buffer: 1 ul <br /> | T4 buffer: 1 ul <br /> | ||
| Line 80: | Line 80: | ||
<br/> | <br/> | ||
| + | |||
====<p style="font-size:15px; background-color:#66bbff;"><b>Sequencing results of pCerulean_Zegfr:1907_MiddleLinker_VP2/3_HSPG-KO</b></p>==== | ====<p style="font-size:15px; background-color:#66bbff;"><b>Sequencing results of pCerulean_Zegfr:1907_MiddleLinker_VP2/3_HSPG-KO</b></p>==== | ||
<b>Investigator: Hanna</b><br> | <b>Investigator: Hanna</b><br> | ||
Revision as of 19:45, 1 October 2010
136. labday 01.10.2010
mini preps of CD clones
Investigator: Kira
c(p689)= 115 ng/ul
c(690)= 151, 20 ng/ul
c(691)= 122,27 ng/ul
c(p692) = 126,42 ng/ul
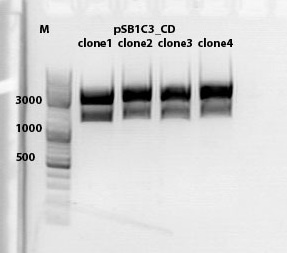
test digestion of CD clones
Investigator: Kira
| Components | sample Volume/µL |
| DNA | 4,0 µl |
| BSA (10x) | 2 µl |
| Buffer no. 4 | 2,0 µl |
| Enzyme 1 XbaI | 1,0 µl |
| Enzyme 2 AgeI | 1,5 µl |
| H2O | 9,5 µl |
| Total volume | 20 |
incubation @ 37 C for approx. 2 h
1% agarose gel
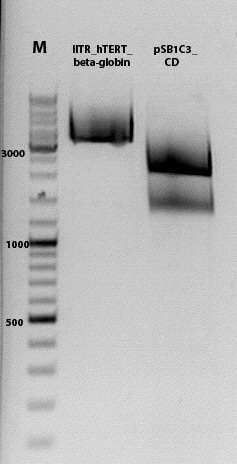
Biobrick assembly pSB1C3_lITR_hTERT_beta-globin_CD
Investigator: Kira
c(pSB1C3_lITR_hTERT_beta-globin)= 333 ng/ul
c(pSB1C3_CD)= 151 ng/ul
| Components | vector Volume/µL | insert Volume/µL |
| DNA | 4,5 µl | 6 |
| BSA (10x) | 3 µl | 3 |
| Buffer no. 4 | 3,0 µl | 3 |
| Enzyme 1 XbaI | 0 µl | 1,5 |
| Enzyme 2 SpeI | 1,5 µl | 0 |
| Enzyme 3 PstI-HF | 1,0 | 1 |
| H2O | 17 | 15,5 |
| Total volume | 25 |
incubation @ 37 C for approx. 2 h
1% agarose gel
Ligation
DNA-mix: 8 ul (vector 4,6ul)+(insert 3,4 ul)
T4 ligase: 1 ul
T4 buffer: 1 ul
Incubation @ RT for 30 min
Transformation was performed according to the standard protocol w BL21 cells.
Sequencing results of pCerulean_Zegfr:1907_MiddleLinker_VP2/3_HSPG-KO
Investigator: Hanna
Comment: All N-terminal fusion approaches with VP2/3_HSPG-KO revealed positive results except of pCerulean_Zegfr:1907_MiddleLinker_VP2/3_HSPG-KO. Another clone was picked, preped, test digested and sent for sequencing.
Conclusion: Sequencing results revealed positive results.
Picking clones of pGA14_MiddleLinker_VP2/3_insCap and pGA14_MiddleLinker_VP2/3_HSPG-KO
Investigator: Hanna
Unfortunately there grew a bacteria lawn over night - it was hardly not possible to pick clones. Nevertheless I tried and picked 2 clones of pGA14_MiddleLinker_VP2/3_insCap and pGA14_MiddleLinker_VP2/3_HSPG-KO.
To do: Mini-Prep and test digestion.
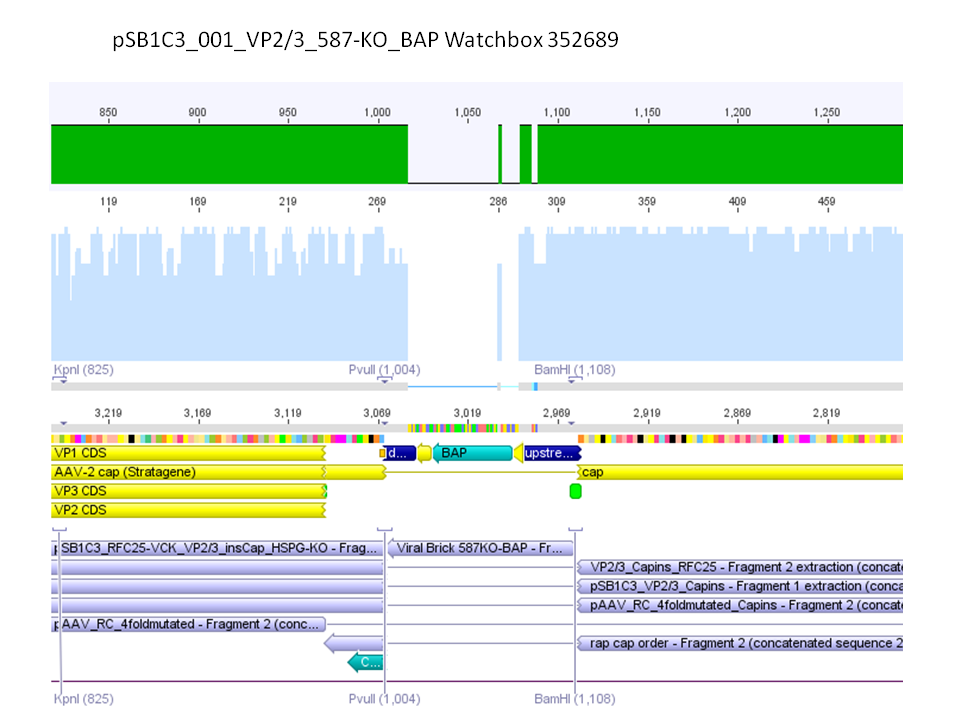
Sequencing results of pSB1C3_001_VP2/3_587-KO_BAP and pSB1C3_001_VP2/3_587-KO_6xHis
Investigator: Hanna
Comment: For the creation of our super constructs, the His-Tag and the BAP motif need to be cloned into VP2/3 for N-terminal fusion to VP2. Sequencing results showed that the 6xHis and BAP motif was not cloned into VP2/3_insCap.
To do: Clone 587-KO_6xHis and 587-KO_BAP into pSB1C3_001_VP2/3_insCap 1. via digestion of inserts and 2. via hybridization of referring oligos - digestion of vector with 800-900 ng.
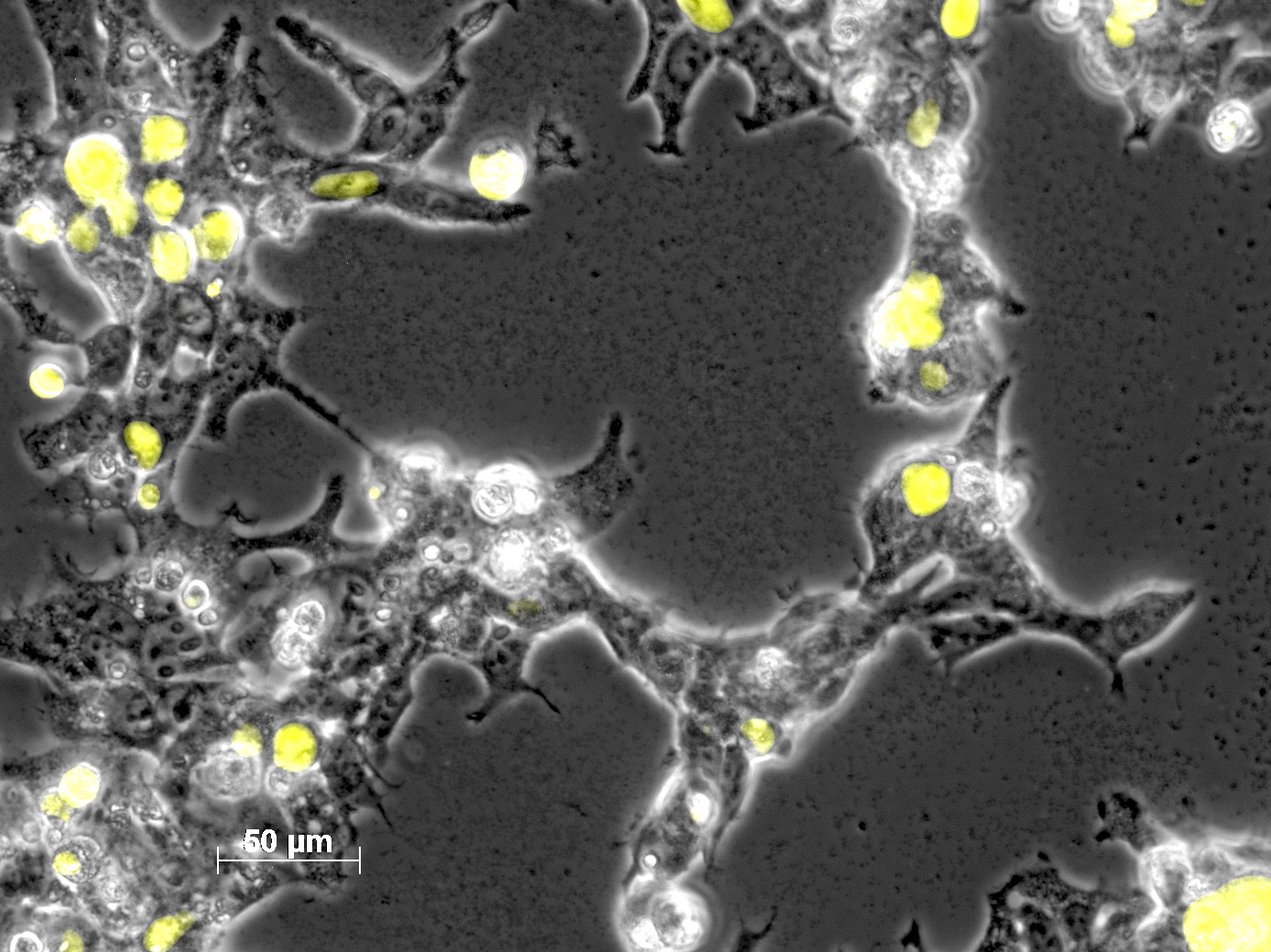
Impressions of transfection of AAV293 with pCerulean_VP1up_NLS_mVenus_VP2/3_insCap
Investigator: Adrian
Yesterday AAV293 cells were transfected with a pCerulean_VP1up_NLS_mVenus_VP2/3_insCap construct. Today, nuclear localization of the produced mVenus_VP2/3_insCap fusion protein was visible and can be nicely seen in the following pictures:
 "
"