Team:Edinburgh/Notebook/Blue light sensor
From 2010.igem.org
| (23 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
h2{ background-color: white; margin: 0 ! important; border-left: 0.5em solid white; border-right: 0.5em solid white} | h2{ background-color: white; margin: 0 ! important; border-left: 0.5em solid white; border-right: 0.5em solid white} | ||
p{ background-color: white; margin: 0; border-left: 0.5em solid white; border-right: 0.5em solid white;} | p{ background-color: white; margin: 0; border-left: 0.5em solid white; border-right: 0.5em solid white;} | ||
| - | ul{ background-color: white; | + | ul{ background-color: white; margin: 0; padding: 1.5em} |
| - | ol{ background-color: white; | + | ol{ background-color: white; margin: 0; padding: 1.5em} |
| + | table{ width: 100%} | ||
| + | #menubar ul{ background: none; margin: 0.3em 0 0 1.5em; padding: 0} | ||
</style> | </style> | ||
</head> | </head> | ||
| Line 41: | Line 43: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Genomic">submitted parts</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Genomic">submitted parts</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Genomic">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Genomic">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Project/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 48: | Line 50: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial" class="dir">bacterial BRIDGEs</a> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial" class="dir">bacterial BRIDGEs</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Core_repressilator">the | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Core_repressilator">the project</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">red light</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">red light</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_sensor">red sensor</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_sensor">red sensor</a></li> | ||
| Line 57: | Line 59: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Bacterial">submitted parts</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Bacterial">submitted parts</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Bacterial">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Bacterial">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 68: | Line 70: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Bacterial">the bacterial model</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Bacterial">the bacterial model</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Signalling">the signalling model</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Signalling">the signalling model</a></li> | ||
| + | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Tools">tools</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Modelling">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Modelling">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 76: | Line 79: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human" class="dir">human BRIDGEs</a> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human" class="dir">human BRIDGEs</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/ | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Communication">communication of science</a></li> |
| + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Branding">iGEM survey</a></li> | ||
| + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Conversations">conversations</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Epic">the epic</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Epic">the epic</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/ | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/FutureApps">future applications</a></li> |
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Human | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Human">further thoughts</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 87: | Line 92: | ||
<ul> | <ul> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Collaboration">collaboration</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Collaboration">collaboration</a></li> | ||
| + | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Attribution">attribution</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/BRIDGE">BRIDGE</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/BRIDGE">BRIDGE</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Red_light_producer">red light</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Red_light_producer">red light</a></li> | ||
| Line 92: | Line 98: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Blue_light_producer">blue light</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Blue_light_producer">blue light</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Blue_light_sensor">blue sensor</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Blue_light_sensor">blue sensor</a></li> | ||
| + | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Green_light_producer">green light</a></li> | ||
| + | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Green_light_sensor">green sensor</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Modelling">modelling</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Modelling">modelling</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Safety">safety</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Notebook/Safety">safety</a></li> | ||
| Line 107: | Line 115: | ||
<br> | <br> | ||
<br> | <br> | ||
| + | |||
| + | <div id="windowbox" style="border: .2em solid #660000; padding: 5px; position:fixed; top:50%; right:30px; width:8%;"> | ||
| + | <span style="color:ivory;">These <b>notes</b> correspond to the part of the <b>project</b> detailed <a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Blue_light_sensor">here</a>.</span> | ||
| + | |||
| + | </div> | ||
</html> | </html> | ||
==Blue Light Sensor== | ==Blue Light Sensor== | ||
| - | + | <br> | |
'''14/07/10''' | '''14/07/10''' | ||
| Line 121: | Line 134: | ||
*BBa_191009 (85 ng/µL) – mutant | *BBa_191009 (85 ng/µL) – mutant | ||
*Sosnick vector containing TrpR (393 ng/µL) – vector | *Sosnick vector containing TrpR (393 ng/µL) – vector | ||
| - | |||
| - | |||
'''16/07/10''' | '''16/07/10''' | ||
| Line 133: | Line 144: | ||
Note: Labeling on tubes is unclear – K191009 and K191004 got mixed up, so plating both tubes on both Kan and amp | Note: Labeling on tubes is unclear – K191009 and K191004 got mixed up, so plating both tubes on both Kan and amp | ||
| - | |||
'''22/07/10''' | '''22/07/10''' | ||
| Line 157: | Line 167: | ||
Lanes- 1- marker, 2-9 sampels as in order, 10-empty, 11, 12- marker instead of the samples- need to run last two again | Lanes- 1- marker, 2-9 sampels as in order, 10-empty, 11, 12- marker instead of the samples- need to run last two again | ||
| - | [[ | + | [[Image:109.JPG]] |
| - | [[ | + | |
| + | [[Image:111.JPG]] | ||
LAST TWO LANES- NSAMPLES NO 13 AND 14 FROM THE ABOVE LIST | LAST TWO LANES- NSAMPLES NO 13 AND 14 FROM THE ABOVE LIST | ||
| - | |||
| - | |||
'''28/07/10''' | '''28/07/10''' | ||
| Line 169: | Line 178: | ||
Trans. 2: BBa_K191004+ K191009- read out + mutated LovTap | Trans. 2: BBa_K191004+ K191009- read out + mutated LovTap | ||
| - | 30/7/10 | + | '''30/7/10''' |
set up experiments to see if the blue light sensor will activate the response system under the daylight. Both liquid cultures and plates were set up, with red and white colonies- first supposed to turn white in the daylight and stay red in the dark (under the foil), second ones- no change of the colour. The same applies to the liquid cultures. However, the experiment failed- no proper growth was observed, and if there was a growth - it was very weak. No pigment observed, apart from the small points in the centre. | set up experiments to see if the blue light sensor will activate the response system under the daylight. Both liquid cultures and plates were set up, with red and white colonies- first supposed to turn white in the daylight and stay red in the dark (under the foil), second ones- no change of the colour. The same applies to the liquid cultures. However, the experiment failed- no proper growth was observed, and if there was a growth - it was very weak. No pigment observed, apart from the small points in the centre. | ||
Hypothesis- not stable plasmids-- cells look like they are not really resistant. Recorded growth: | Hypothesis- not stable plasmids-- cells look like they are not really resistant. Recorded growth: | ||
| Line 192: | Line 201: | ||
|} | |} | ||
| + | '''2/8/10''' | ||
| + | Run the digest of the minipreps from the white and red cultures of double transformants. No proper DNA band in the solution- repeat the procedure or not? Don't know what to do.... again. | ||
| - | + | [[Image:2.08.JPG]] | |
| - | |||
| - | |||
Collect the thoughts and ideas and start again tomorrow! | Collect the thoughts and ideas and start again tomorrow! | ||
| Line 214: | Line 223: | ||
by Chairman-->' If we can't find this information, I think our best course is to | by Chairman-->' If we can't find this information, I think our best course is to | ||
PCR out both inserts and combine them in a single vector, preferably pSB1C3.' | PCR out both inserts and combine them in a single vector, preferably pSB1C3.' | ||
| - | |||
'''9/8/2010''' | '''9/8/2010''' | ||
| Line 255: | Line 263: | ||
* BAD NEWS: there is a suspicion of 1bp deletion in LovTap!!! Chairman noticed it when he was checking the parts base by base. Should have done it much much earlier probably to save the time... | * BAD NEWS: there is a suspicion of 1bp deletion in LovTap!!! Chairman noticed it when he was checking the parts base by base. Should have done it much much earlier probably to save the time... | ||
* BAD NEWS: lack of the proper thinking put in doubt all the results from previous two experiments (that is why you can't find any files of it attached)- there was no IPTG added to the samples, therefore LOVTap expression was not induced- no point to do T-test right now... :( Well, at least I will learn from it and add it next time! | * BAD NEWS: lack of the proper thinking put in doubt all the results from previous two experiments (that is why you can't find any files of it attached)- there was no IPTG added to the samples, therefore LOVTap expression was not induced- no point to do T-test right now... :( Well, at least I will learn from it and add it next time! | ||
| - | |||
'''18/08/10''' | '''18/08/10''' | ||
| - | * tested LovTap under the blue light | + | * tested LovTap under the blue light (blue LEDs, 470 nm wavelength, 2200 mcd/B, V=4.5 V; RS 466-3548). |
* conditions: | * conditions: | ||
Liquid cultures (same as for minipreps) were grown overnight in 37C with antibiotics. | Liquid cultures (same as for minipreps) were grown overnight in 37C with antibiotics. | ||
The next day, bottles with 3.5 ml of broth were inoculated with the 0.5 ml of appropriate cultures (0.5. ml of broth added as a control). | The next day, bottles with 3.5 ml of broth were inoculated with the 0.5 ml of appropriate cultures (0.5. ml of broth added as a control). | ||
| - | *sample designation: (# refers to the clone number form the original plate). Original plates are the ones made on | + | *sample designation: (# refers to the clone number form the original plate). Original plates are the ones made on 25/7 and have 2 clones on each plates (clones1-3 are K191003&4 double transformants, clones 4-6 are K191009+4 double transformants). |
# K191003+4 (#1) + Amp + Kan + IPTG + LIGHT | # K191003+4 (#1) + Amp + Kan + IPTG + LIGHT | ||
| Line 279: | Line 286: | ||
* samples designated DARK were wrapped in the aluminium foil straight after the inoculation; the cap was not wrapped (not see through anyway)so that samples could be easily taken without unwrapping the bottles from the foil. | * samples designated DARK were wrapped in the aluminium foil straight after the inoculation; the cap was not wrapped (not see through anyway)so that samples could be easily taken without unwrapping the bottles from the foil. | ||
*All the samples were then taken to the shaker in the hot room and placed in prepared spots: for the dark- anywhere on an easily accessible holder, for light- in 2 rows of holders. | *All the samples were then taken to the shaker in the hot room and placed in prepared spots: for the dark- anywhere on an easily accessible holder, for light- in 2 rows of holders. | ||
| - | *Light samples were covered with the cardboard box with blue LEDs attached on the sides, in a way to optimize light accession to the samples | + | *Light samples were covered with the cardboard box with blue LEDs attached on the sides, in a way to optimize light accession to the samples. Box was securely taped to the shaker. |
* Measurments were taken every half an hour: 200ul of the samples were placed in the numbered cuvette and then 800ul of water was added. Readings of fluroscence (green mode) were taken in standard fluorescence units (SFU, two readings each). Then optical density was measured (upstairs in the incubator room) at 360 nm wavelength. Control was used to calibrate the machine (set ref button). Also 2 readings were taken. Samples were shaken by inversion prior to taking measurment. | * Measurments were taken every half an hour: 200ul of the samples were placed in the numbered cuvette and then 800ul of water was added. Readings of fluroscence (green mode) were taken in standard fluorescence units (SFU, two readings each). Then optical density was measured (upstairs in the incubator room) at 360 nm wavelength. Control was used to calibrate the machine (set ref button). Also 2 readings were taken. Samples were shaken by inversion prior to taking measurment. | ||
* Experiment was stopped after 3 hours (6 sets of reading taken). | * Experiment was stopped after 3 hours (6 sets of reading taken). | ||
*'''Result analysis''' | *'''Result analysis''' | ||
| - | |||
| - | [[ | + | [[Image:Lt18aug.JPG]] |
To generate the data for the graph: | To generate the data for the graph: | ||
| Line 294: | Line 300: | ||
# graph was made with series named the same as the samples | # graph was made with series named the same as the samples | ||
| - | |||
**BAD BAD VERY BAD NEWS: after 2 hours of measurments the results of the sequencing were know and revealed that THERE IS A 1BP DELETION IN LOVTAP 03 AND 09 WHICH RESULTS IN FRAMESHIFT AND LACK OF THE FUNCTIONAL PRODUCT!!!! :(:(:( despite this information, it was decided that the experiment will be finished and analysed as initially planned. | **BAD BAD VERY BAD NEWS: after 2 hours of measurments the results of the sequencing were know and revealed that THERE IS A 1BP DELETION IN LOVTAP 03 AND 09 WHICH RESULTS IN FRAMESHIFT AND LACK OF THE FUNCTIONAL PRODUCT!!!! :(:(:( despite this information, it was decided that the experiment will be finished and analysed as initially planned. | ||
| Line 308: | Line 313: | ||
* Self -ligation overnight (MABEL self ligation- 12ul of H2O, 4ul of DNA, 2 ul of Ligase buffer, 1 ul of T4 DNA ligase, 1ul of T4 polynucleotide kinase. | * Self -ligation overnight (MABEL self ligation- 12ul of H2O, 4ul of DNA, 2 ul of Ligase buffer, 1 ul of T4 DNA ligase, 1ul of T4 polynucleotide kinase. | ||
* transformation and setting up the liquid cultures for the minipreps over the weekend. | * transformation and setting up the liquid cultures for the minipreps over the weekend. | ||
| + | |||
| + | [[Image:123.JPG]] | ||
'''23/08/2010''' | '''23/08/2010''' | ||
* mini-prep DNA from the colonies after PCR- marker worn off in the shaker,so 3.2. and 9.1. got mixed up- probably sequencing necessary to distinguish the mutated version from non-mutated | * mini-prep DNA from the colonies after PCR- marker worn off in the shaker,so 3.2. and 9.1. got mixed up- probably sequencing necessary to distinguish the mutated version from non-mutated | ||
| - | * digest minipreps with EcoRI, run the gel | + | * digest minipreps with EcoRI, run the gel[photo] |
* run 5ul of the ligated 03 and 09 on the gel /also Richards samples/. | * run 5ul of the ligated 03 and 09 on the gel /also Richards samples/. | ||
| + | |||
| + | [[Image:124.JPG]] | ||
'''24/08/2010''' | '''24/08/2010''' | ||
| Line 318: | Line 327: | ||
* minipreped 6 more clones of mutated LovTap 03 and 09 from the same plate, labelled- 3.6, 3.7, 3.8, 9.4, 9.5, 9.6. Run the digest with EcoRI. Note: 3.7 sample is higly diluted as 80ul of EB was used instead of 40ul. oops. | * minipreped 6 more clones of mutated LovTap 03 and 09 from the same plate, labelled- 3.6, 3.7, 3.8, 9.4, 9.5, 9.6. Run the digest with EcoRI. Note: 3.7 sample is higly diluted as 80ul of EB was used instead of 40ul. oops. | ||
* delivered mutated 3.? and 9.2 minipreped clones for sequencing /only 2 reactions: each sample- forward/. | * delivered mutated 3.? and 9.2 minipreped clones for sequencing /only 2 reactions: each sample- forward/. | ||
| + | |||
| + | [[Image:LT100.jpg]] | ||
| + | |||
| + | '''25/9/2010''' | ||
| + | * trpR mutant received form KEIO collection- generously privided by Lucas Black (The University of Edinburgh). | ||
| + | '''1/10/2010''' | ||
| + | * LOVTap-RFP in JM109 | ||
'''04/10/10''' | '''04/10/10''' | ||
We may have a working LovTap construct + red/white (RFP) reporter system thanks to Mexico. | We may have a working LovTap construct + red/white (RFP) reporter system thanks to Mexico. | ||
| - | + | * Cutting Mexico's LOVTap with EcoRi/PstI & transfer into pSB1C3 plasmid (Part 1) | |
| + | * Adding reporter- cut out with XbaI/PstI from K191004 part provided by Lausanne (PArt 2) | ||
| + | * Ligation of Part 1 and Part 2., selection for red colonies. | ||
Minor characterisation experiment:<br> | Minor characterisation experiment:<br> | ||
| Line 329: | Line 347: | ||
-One plate left in the light, the other left in the dark<br> | -One plate left in the light, the other left in the dark<br> | ||
-If the one in the light grows red and the one in the dark doesn't, it is a good indication that the construct is working<br><br> | -If the one in the light grows red and the one in the dark doesn't, it is a good indication that the construct is working<br><br> | ||
| + | |||
| + | '''5/10/2010''' | ||
| + | * transforming trpR mutant with LOVTap- RFP plasmid. | ||
| + | |||
| + | '''8/10/2010''' | ||
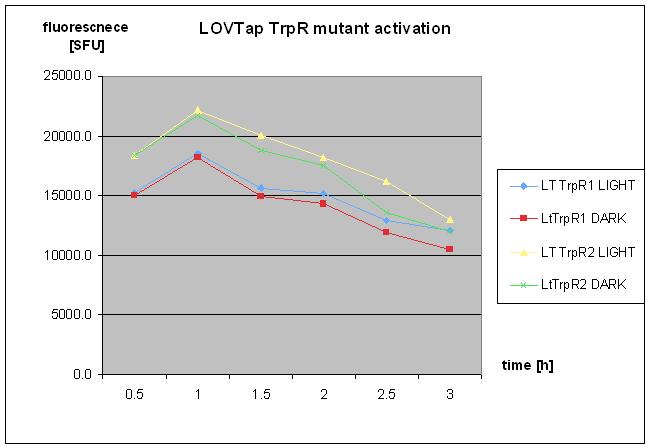
| + | * repetaed LOVTap charactierzation- protocol followed as on 18th of August 2010- with TrpR mutant and WT under light/dark conidtion. | ||
| + | * no clear response was seen. | ||
| + | |||
| + | [[Image:8octLovtap.jpg]] | ||
| + | |||
| + | [[Image:8octTRPR.jpg]] | ||
| + | |||
| + | '''11/10/2010''' | ||
| + | * Transforming plasmid Ptrp with RFP reporter to JM109 strian- control for further experiments. | ||
| + | |||
| + | '''14/10/2010''' | ||
| + | * PRi in trpR host- control plasmid | ||
| + | |||
| + | '''22/10/2010''' | ||
| + | * checking temperature response of LOVTap | ||
| + | * setting up samples with CMM minum medium, enriched in thymine, trace elements and glucose--> for TrpR mutant. | ||
| + | CELLS FAIL TO GROW!:( | ||
Latest revision as of 02:12, 28 October 2010
Blue Light Sensor
14/07/10
Received parts from Lausanne:
- BBa_191006 (113 ng/µL) – LovTAP alone
- BBa_191004 (166 ng/µL) – Readout
- BBa_191003 (230 ng/µL) – whole system
- BBa_191009 (85 ng/µL) – mutant
- Sosnick vector containing TrpR (393 ng/µL) – vector
16/07/10
Transformations using 100 µL of cells, 900 µL broth
- K191004 - amp
- K191003 - kanamycin
- K191009 – kanamycin
- Sosnick vector with TrpR – amp
Note: Labeling on tubes is unclear – K191009 and K191004 got mixed up, so plating both tubes on both Kan and amp
22/07/10
- Minipreps: K0998010 – 5x (1 didn’t grow, but did anyway)
- Blue sensors (3 biobricks and Sosnick vector) – cultures set to grow overnight
25/07/10 Digest of the blue light sensor minipreps was performed with EcoRI and BamHI; samples:
- 3- K191004; 900ul, (3)
- 4- K191004; 100ul, (2)
- 5-K191009; 100ul, (1)
- 6- K191009; 100ul, (6)
- 7- K191009; 900ul, (4)
- 8- K191009; 100ul, (2)
- 9- K191003; 900ul, (2)
- 10- K191003; 900ul, (5)
- 11- K191003; 100ul, (4)
- 12- K191003; 100ul, (1)
- 13- K191004; 100ul, (3)
- 14- K191004; 900ul, (5)
Lanes- 1- marker, 2-9 sampels as in order, 10-empty, 11, 12- marker instead of the samples- need to run last two again

28/07/10
Double transformants made using a novel cowboy procedure- double transformation with 4ul of each piece od DNA. Chances of succes- very very low... But at least it will be proved that it won't work! Trans. 1: BBa_K191004+ K191003- read out + LovTap Trans. 2: BBa_K191004+ K191009- read out + mutated LovTap
30/7/10 set up experiments to see if the blue light sensor will activate the response system under the daylight. Both liquid cultures and plates were set up, with red and white colonies- first supposed to turn white in the daylight and stay red in the dark (under the foil), second ones- no change of the colour. The same applies to the liquid cultures. However, the experiment failed- no proper growth was observed, and if there was a growth - it was very weak. No pigment observed, apart from the small points in the centre. Hypothesis- not stable plasmids-- cells look like they are not really resistant. Recorded growth:
| K191004+9 control | K191004+9 | K191004+3 control | K191004+3 | |
| DARK | ++ , white | +++ , white | ++ , white | +++ , white with red points |
| LIGHT | + , white | +++ , white with red point | + -almost no growth | ++ white |
2/8/10
Run the digest of the minipreps from the white and red cultures of double transformants. No proper DNA band in the solution- repeat the procedure or not? Don't know what to do.... again.
Collect the thoughts and ideas and start again tomorrow!
6/8/10
double transformants were prepared again, using non-cowboy procedure. 04 cells were first MADE COMPETENT, then transformed with 03 and 09, also 1 neg control was plated. Grow white on the intitial plate, then turned red if subcultured.
7-8/08
'Subbed these to duplicate fresh plates and incubated overnight at room temperature either under a fluorescent lamp or wrapped in foil - all white in both cases (and growth not great, probably because of low temperature)'. The plates were moved to 37C with the lamp /one under the light, one in the dark. so far -no results, checking in an hour/.
Another idea- check for stability of the insert- we don't know if the plasmids are compatibile. Streaking from original plates to check if single colonies will be a mixture of white and red or just red- origin? no plasmid maps from Lausanne. by Chairman-->' If we can't find this information, I think our best course is to PCR out both inserts and combine them in a single vector, preferably pSB1C3.'
9/8/2010
to do:
- primers for the parts to put them in a vector
- check the lamp every hour!!
go thruough Lausanne website to find the plasmid maps get back to Mexico about the parts and etc. find out the conditions for the blue light sensor assay!
10/08/10
- assay with dark/light liquid cultures - fluorescence measurment and OD, after 5 h in the shaker under lamp- see video and pictures
- email to Mexico
11/08/10
- primer designed for combining LOVTap and Readout1 into 1 cell
- repeated assay with the smae samples, after overngiht in RT and %5h under lamp (for the light samples) and RT without shaking
- primary cultures streaked to amp/kan/IPTG plates- part 4+9 and 4+3 -three clones each
12/08/10
- results analysis after LOVTap experiment:check file, add graphs later.
- email from Mexico about Blue promoter
16/08/10
- streaked single clone for 9+4 and 3+4 (clones number 5 and 2, respectively)- for tomorrow's experiment
- minipreped mysterious double transformants and Sacb smples
- tried out the box on the shaker- unscrewed the things.
- made the graphs from the second reading
- 03 and 09 LovTap taken for sequencing.
to do: decide on the illumination length and think about controls and how many samples do you want to put. check Lausanne website t-test to check to importancy of the differences between the means.
17/07/10
- desinged experimental set up for the LovTap blue light response experiment /see next day/
- BAD NEWS: there is a suspicion of 1bp deletion in LovTap!!! Chairman noticed it when he was checking the parts base by base. Should have done it much much earlier probably to save the time...
- BAD NEWS: lack of the proper thinking put in doubt all the results from previous two experiments (that is why you can't find any files of it attached)- there was no IPTG added to the samples, therefore LOVTap expression was not induced- no point to do T-test right now... :( Well, at least I will learn from it and add it next time!
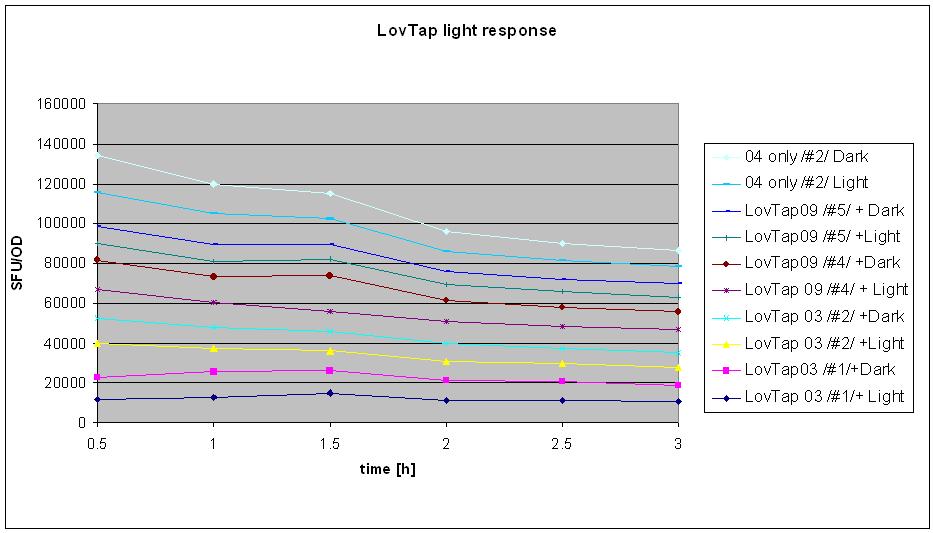
18/08/10
- tested LovTap under the blue light (blue LEDs, 470 nm wavelength, 2200 mcd/B, V=4.5 V; RS 466-3548).
- conditions:
Liquid cultures (same as for minipreps) were grown overnight in 37C with antibiotics. The next day, bottles with 3.5 ml of broth were inoculated with the 0.5 ml of appropriate cultures (0.5. ml of broth added as a control).
- sample designation: (# refers to the clone number form the original plate). Original plates are the ones made on 25/7 and have 2 clones on each plates (clones1-3 are K191003&4 double transformants, clones 4-6 are K191009+4 double transformants).
- K191003+4 (#1) + Amp + Kan + IPTG + LIGHT
- K191003+4 (#1) + Amp + Kan IPTG + DARK
- K191003+4 (#2) + Amp + Kan IPTG + LIGHT
- K191003+4 (#2) + Amp + Kan IPTG + DARK
- K191009+4 (#4) + Amp + Kan IPTG +LIGHT
- K191009+4 (#4) + Amp + Kan IPTG +DARK
- K191009+4 (#5) + Amp + Kan IPTG +LIGHT
- K191009+4 (#5) + Amp + Kan IPTG +DARK
- K191004 ONLY #2+ Amp + LIGHT
- K191004 ONLY #2+ Amp + DARK
- neg. control- broth, Amp + Kan + IPTG + LIGHT
- samples designated DARK were wrapped in the aluminium foil straight after the inoculation; the cap was not wrapped (not see through anyway)so that samples could be easily taken without unwrapping the bottles from the foil.
- All the samples were then taken to the shaker in the hot room and placed in prepared spots: for the dark- anywhere on an easily accessible holder, for light- in 2 rows of holders.
- Light samples were covered with the cardboard box with blue LEDs attached on the sides, in a way to optimize light accession to the samples. Box was securely taped to the shaker.
- Measurments were taken every half an hour: 200ul of the samples were placed in the numbered cuvette and then 800ul of water was added. Readings of fluroscence (green mode) were taken in standard fluorescence units (SFU, two readings each). Then optical density was measured (upstairs in the incubator room) at 360 nm wavelength. Control was used to calibrate the machine (set ref button). Also 2 readings were taken. Samples were shaken by inversion prior to taking measurment.
- Experiment was stopped after 3 hours (6 sets of reading taken).
- Result analysis
To generate the data for the graph:
- means were calculated from 2 readings of optical density and fluorescence
- (mean SFU minus background SFU) was divided by (mean OD minus background OD, if different from 0).
- table was made for the samples at different points of time- e.g. sample 1 after 30 min, 60 min, 90 min, 120 min etc.
- graph was made with series named the same as the samples
- BAD BAD VERY BAD NEWS: after 2 hours of measurments the results of the sequencing were know and revealed that THERE IS A 1BP DELETION IN LOVTAP 03 AND 09 WHICH RESULTS IN FRAMESHIFT AND LACK OF THE FUNCTIONAL PRODUCT!!!! :(:(:( despite this information, it was decided that the experiment will be finished and analysed as initially planned.
19/08/2010
- Pcr- mutagenensis of LovTap- Mabel, adding missing bp.
- Puryfing PCR product from the solution.
20/8/2010
- PCR run on the gel -correct band of ~5.5. kb size, suggest that the PCR worked well. couple of other fainter bands
- Self -ligation overnight (MABEL self ligation- 12ul of H2O, 4ul of DNA, 2 ul of Ligase buffer, 1 ul of T4 DNA ligase, 1ul of T4 polynucleotide kinase.
- transformation and setting up the liquid cultures for the minipreps over the weekend.
23/08/2010
- mini-prep DNA from the colonies after PCR- marker worn off in the shaker,so 3.2. and 9.1. got mixed up- probably sequencing necessary to distinguish the mutated version from non-mutated
- digest minipreps with EcoRI, run the gel[photo]
- run 5ul of the ligated 03 and 09 on the gel /also Richards samples/.
24/08/2010
- repeated digest of mutated LovTap samples: 3.1, 3.?, 3.3., 3.4 9.?, 9.2 with EcoRi
- minipreped 6 more clones of mutated LovTap 03 and 09 from the same plate, labelled- 3.6, 3.7, 3.8, 9.4, 9.5, 9.6. Run the digest with EcoRI. Note: 3.7 sample is higly diluted as 80ul of EB was used instead of 40ul. oops.
- delivered mutated 3.? and 9.2 minipreped clones for sequencing /only 2 reactions: each sample- forward/.
25/9/2010
- trpR mutant received form KEIO collection- generously privided by Lucas Black (The University of Edinburgh).
1/10/2010
- LOVTap-RFP in JM109
04/10/10 We may have a working LovTap construct + red/white (RFP) reporter system thanks to Mexico.
- Cutting Mexico's LOVTap with EcoRi/PstI & transfer into pSB1C3 plasmid (Part 1)
- Adding reporter- cut out with XbaI/PstI from K191004 part provided by Lausanne (PArt 2)
- Ligation of Part 1 and Part 2., selection for red colonies.
Minor characterisation experiment:
-LovTap constructed with red-white RFP reporter
-If activated, the blue light sensor transformants should produce red colonies
-Blue light sensor containing-colonies from transformant plate streaked onto two plates
-One plate left in the light, the other left in the dark
-If the one in the light grows red and the one in the dark doesn't, it is a good indication that the construct is working
5/10/2010
- transforming trpR mutant with LOVTap- RFP plasmid.
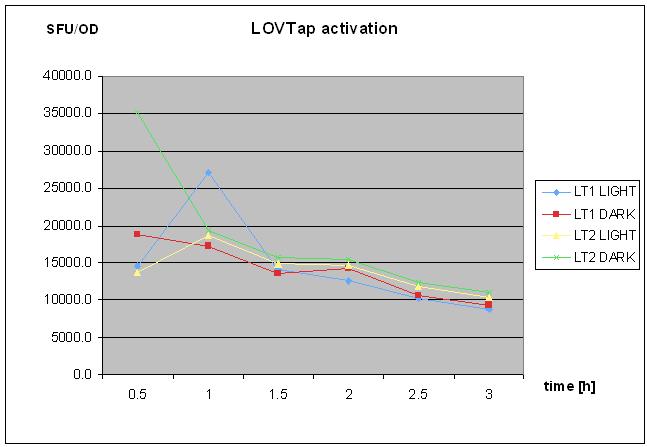
8/10/2010
- repetaed LOVTap charactierzation- protocol followed as on 18th of August 2010- with TrpR mutant and WT under light/dark conidtion.
- no clear response was seen.
11/10/2010
- Transforming plasmid Ptrp with RFP reporter to JM109 strian- control for further experiments.
14/10/2010
- PRi in trpR host- control plasmid
22/10/2010
- checking temperature response of LOVTap
- setting up samples with CMM minum medium, enriched in thymine, trace elements and glucose--> for TrpR mutant.
CELLS FAIL TO GROW!:(
 "
"