Team:ETHZ Basel/Project/Movie
From 2010.igem.org
| Line 1: | Line 1: | ||
{{ETHZ_Basel10}} | {{ETHZ_Basel10}} | ||
| - | {{ | + | {{ETHZ_Basel10_Start}} |
= Project Overview = | = Project Overview = | ||
Revision as of 11:02, 7 October 2010
Project Overview
Molecular Mechanism
Controlling
E. lemming aims to modify the chemotaxis property of E.coli such that, instead of response to a chemical attractant or repellent, the bacterium responds to a light stimulus. Furthermore, this light sensitivity is used to control E.coli’s movement by deciding, at any given time, which type of motion the bacterium will adopt (tumbling or straight run). This leads to a controllable E.coli, which can move to a defined direction, as a result of the combination of tumbling and straight run. The bacteria in the experiment are imaged and, by image processing, the position of a single tracked cell is inferred. By activating a light switch, the user decides whether the bacterium should continue running or should change direction.
Idea
The core idea of our project is to control chemotaxis of E. coli by means of light! We'll realize this by hijacking and perturbing the tumbling / directed flagellar movement apparatus. By coupling directed flagellar movement regulating proteins to a novel synthetic light-sensitive spatial localization system, their activity can be controlled reversibly. A light-sensitive dimerizing complex fused to this regulating proteins at a spatially fixed location is induced by light pulses and therefore localization of the two molecules can be manipulated. Tumbling / directed flagellar movement rates are monitored by image processing algorithms, which are linked to the light-pulse generator. This means that E. coli tumbling is induced or suppressed simply by pressing a light switch! This synthetic network enables control of single E. coli cells: We'll make them move like mindless "Lemmings" in the direction they are forced to go!
Strategy
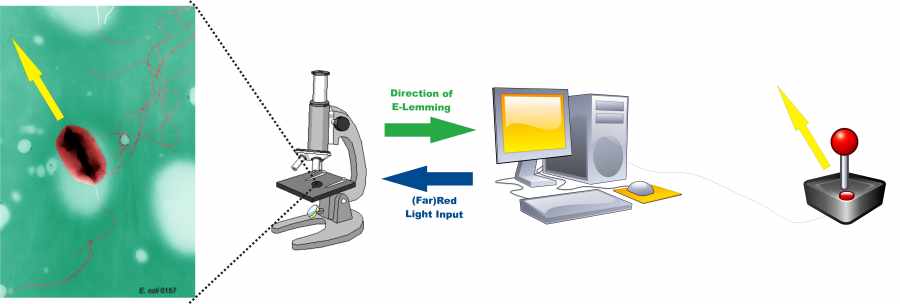
Information processing on the basis of the E. lemming
Inspired by the video game lemmings we want to navigate an E. coli bacterium across the perilous downs of a micro fluidic chamber. The player observes his bacterium on the screen of a microscope and directs it into any desired direction using a joystick.
Our project is divided in two important parts: creating the E. lemming and establishing controller software as an interface between player and microscope.
The E. lemming relies on a modified chemotactic pathway, which is tunable by light pulses. Mathematical models predicted the best layout of the pathway which was then implemented into E. coli. To learn more about the E. lemming see the E. lemming section.
The controller software assesses the position and the direction the bacterium swims to and processes the input of the player into appropriate light pulses. To learn more about controller software please see the Information Processing section.
 "
"


