Team:ETHZ Basel/Modeling/Light Switch
From 2010.igem.org
(→Modeling of the light switch: Archeal light receptor) |
(→Interface to the chemotaxis model) |
||
| Line 16: | Line 16: | ||
The following assumptions have been made according to the [[Team:ETHZ_Basel/Biology/Molecular_Mechanism| molecular mechanism]] to link the light switch and chemotaxis models: | The following assumptions have been made according to the [[Team:ETHZ_Basel/Biology/Molecular_Mechanism| molecular mechanism]] to link the light switch and chemotaxis models: | ||
| - | * Upon red light pulse induction, the two light-sensitive | + | * Upon red light pulse induction, the two light-sensitive protein (LSP1 and LSP2) domains can hetero-dimerize. |
| - | ** CheR is not able to methylate the MCPs anymore, | + | * The TetR-LSP2 can bind to the operator (tetO) on the DNA. |
| - | ** CheY can't be phosphorylated and interact with the motor anymore; nevertheless, it still can be dephosphorylated. | + | * Only after both reactions took place, we assume the Che protein to be spatially dislocated. This means |
| - | + | ** CheR-LSP1 is not able to methylate the MCPs anymore, | |
| - | + | ** CheY-LSP1 can't be phosphorylated and interact with the motor anymore; nevertheless, it still can be dephosphorylated. | |
| - | ** CheB is not able to demethylate the MCPs and can't be phosphorylated anymore, but still can be dephosphorylated, | + | ** CheB-LSP1 is not able to demethylate the MCPs and can't be phosphorylated anymore, but still can be dephosphorylated, |
| - | ** CheZ can't dephosphorylate CheY anymore (This assumption is very unsteady, since CheY is not strictly located). | + | ** CheZ-LSP1 can't dephosphorylate CheY anymore (This assumption is very unsteady, since CheY is not strictly located). |
| + | * If only one of the reactions took place, we assume that the localization and activity of the fusion proteins are the same as for wild-type cells. | ||
All of these assumptions will lead to a decrease of tumbling / directed movement ratio upon red light induction and an increase of corresponding far-red light induction. | All of these assumptions will lead to a decrease of tumbling / directed movement ratio upon red light induction and an increase of corresponding far-red light induction. | ||
Revision as of 14:44, 27 October 2010
The light switch
Modeling of the PhyB/PIF3 light switch
Background
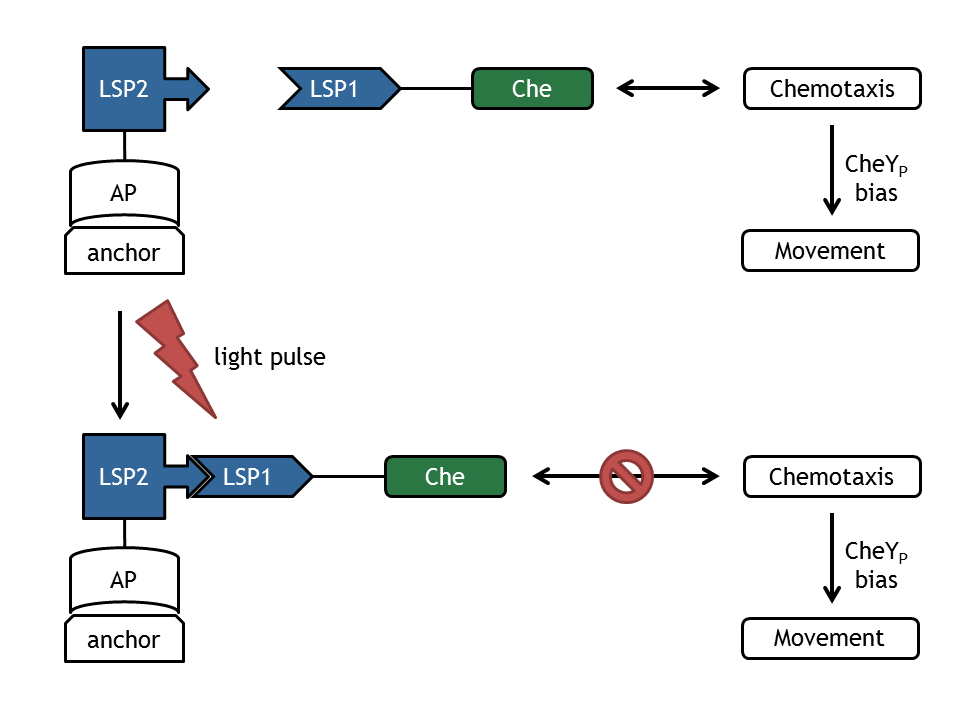
In our biological setup, the relocation of one of the chemotaxis pathway proteins (either CheR, CheB, CheY or CheZ) is achieved by fusing them to a light-sensitive protein LSP1 (either to PhyB or to PIF3), which dimerizes by red light induction with the corresponding LSP2 (PIF3 or PhyB), fused to a spatially dislocated anchor. Since we decided to implement two different models of the chemotaxis pathway (see section Chemotaxis Pathway), modeling all setups implemented in the wet-lab would have resulted in 16 different models:
|{CheR, CheB, CheY, CheZ} × {PhyB, PIF3} × {Model 1, Model 2}|=16.
Inspired by the modular approach used for the Cloning Strategy in the wet-lab, we decided to decrease the combinatorial explosion by also applying a novel modular approach. Not only did this approach reduce the amount of models by a factor of four, it also allowed to widely separate the differential equations of the chemotaxis pathway from those of the light-induced localization system by simultaneously decreasing redundancies and thus decreasing the danger of slip of the pens. The underlying mathematical model for the light-induced relocation was completely developed by us (for a short discussion of the recently by Sorokina et al. published light-induced relocation system and why we did not use it, see here) as well as - to our knowledge - the approach to couple this model to models of the chemotaxis pathway.
Interface to the chemotaxis model
The following assumptions have been made according to the molecular mechanism to link the light switch and chemotaxis models:
- Upon red light pulse induction, the two light-sensitive protein (LSP1 and LSP2) domains can hetero-dimerize.
- The TetR-LSP2 can bind to the operator (tetO) on the DNA.
- Only after both reactions took place, we assume the Che protein to be spatially dislocated. This means
- CheR-LSP1 is not able to methylate the MCPs anymore,
- CheY-LSP1 can't be phosphorylated and interact with the motor anymore; nevertheless, it still can be dephosphorylated.
- CheB-LSP1 is not able to demethylate the MCPs and can't be phosphorylated anymore, but still can be dephosphorylated,
- CheZ-LSP1 can't dephosphorylate CheY anymore (This assumption is very unsteady, since CheY is not strictly located).
- If only one of the reactions took place, we assume that the localization and activity of the fusion proteins are the same as for wild-type cells.
All of these assumptions will lead to a decrease of tumbling / directed movement ratio upon red light induction and an increase of corresponding far-red light induction.
Facing the Combinatorial Explosion
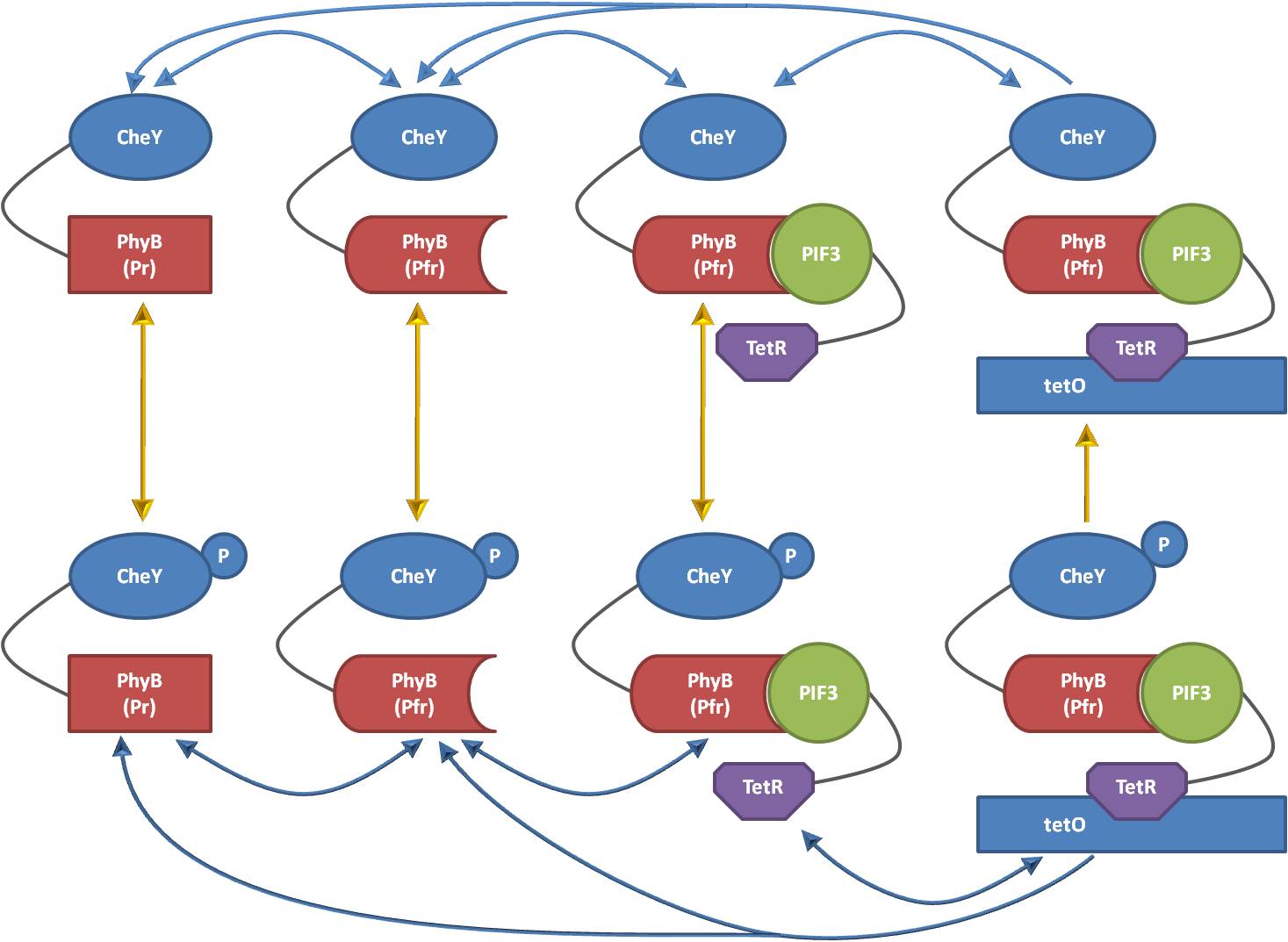
The main problem in separating in silico the chemotaxis pathway from the light-induced relocation system is the non-existence of a hierarchical relationship between the two sub-models: The properties of the chemotaxis pathway are clearly influenced if the active concentration of one of its key species drops, such that giving the amount of localized proteins, obtained by the localization model, as an input to the chemotaxis model is a natural conclusion. However, also the concentration of the localized species of the localization model change depending on the reactions in the chemotaxis model, since several of the Che proteins are modeled as two or more molecular species. The CheY protein for example is modeled as two species, one representing the phosphorylated and one the non-phosphorylated molecular concentration. Thus, the two models cannot be represented as e.g. a hierarchical block structure, which is making a modular approach significant more complicated.
Our approach to modularize the model is based on an observation of the reaction directions which can take place in the overall model. For example, Figure 2 shows all eight species which have to be implemented to adequately describe the localization and phosphorylation states of CheY when fused to PhyB. As can be seen, the reactions of the chemotaxis pathway and the reactions of the light model have different directions. The reactions of the light model are horizontal and independent of the phosphorylation level of CheY. On the other hand, the reactions of the chemotaxis model are vertically. However, one of the reactions is not independent of the states mainly influenced by the light model: When CheY is localized at the DNA, we assume that it is not possible anymore to
Archeal light receptor
Download
The light switch model is included within the Matlab Toolbox and can be downloaded there.
 "
"