Team:ETHZ Basel/Modeling/Interworking
From 2010.igem.org
(→Results) |
|||
| Line 56: | Line 56: | ||
{| border="0" align="right" | {| border="0" align="right" | ||
|- valign="top" | |- valign="top" | ||
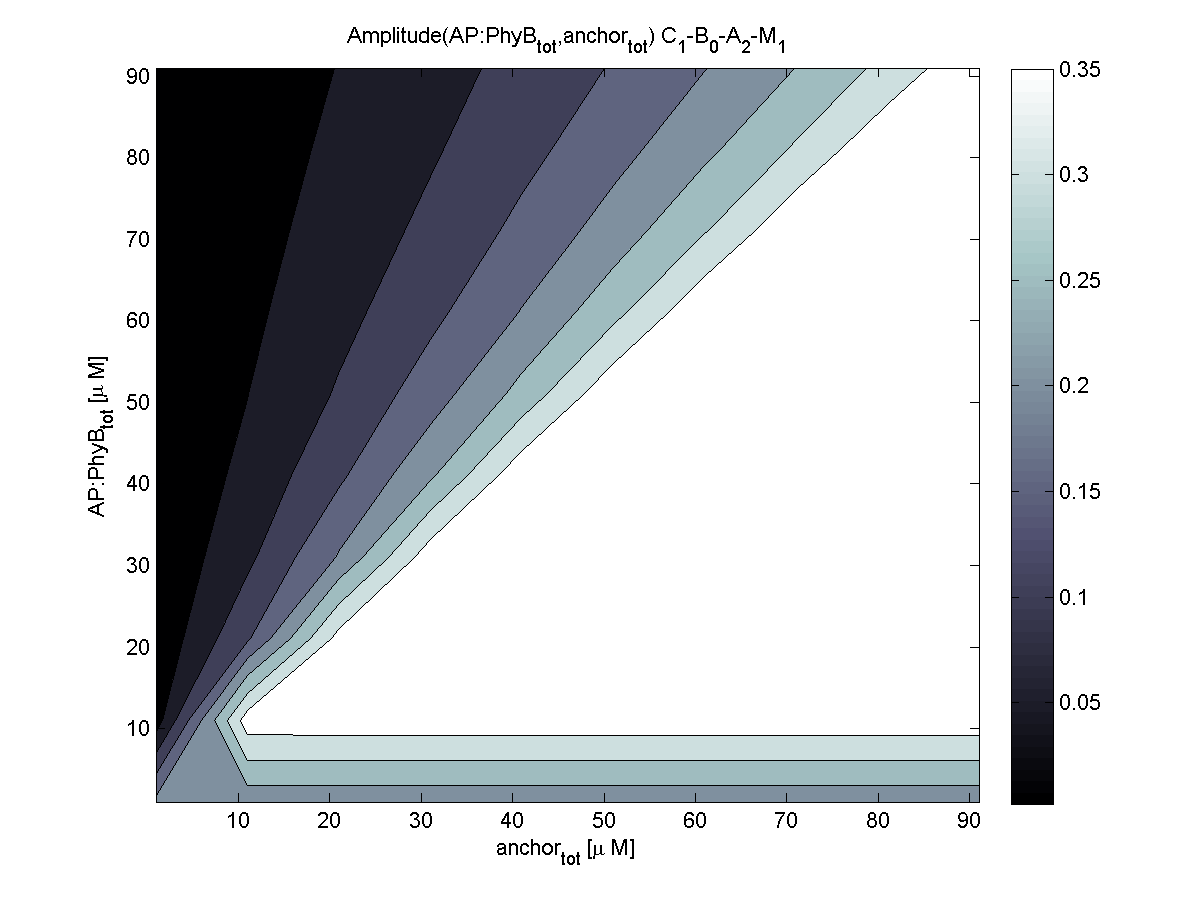
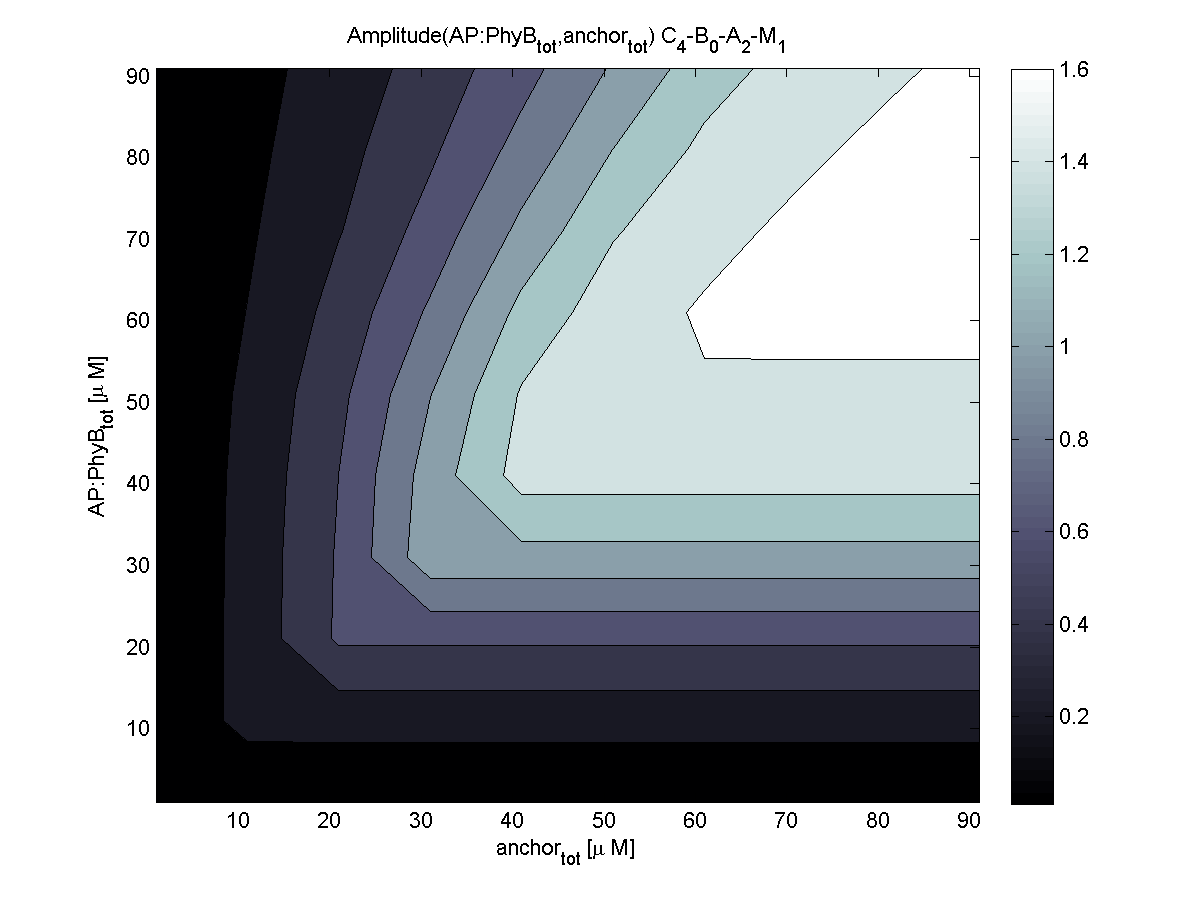
| - | |[[Image:ETHZ_Basel_models_evaluator_phyb.png|thumb|200px|''' | + | |[[Image:ETHZ_Basel_models_evaluator_phyb.png|thumb|200px|'''LSP? = PhyB.''' PhyB coupling is very robust and results in maximal plateau surface.]] |
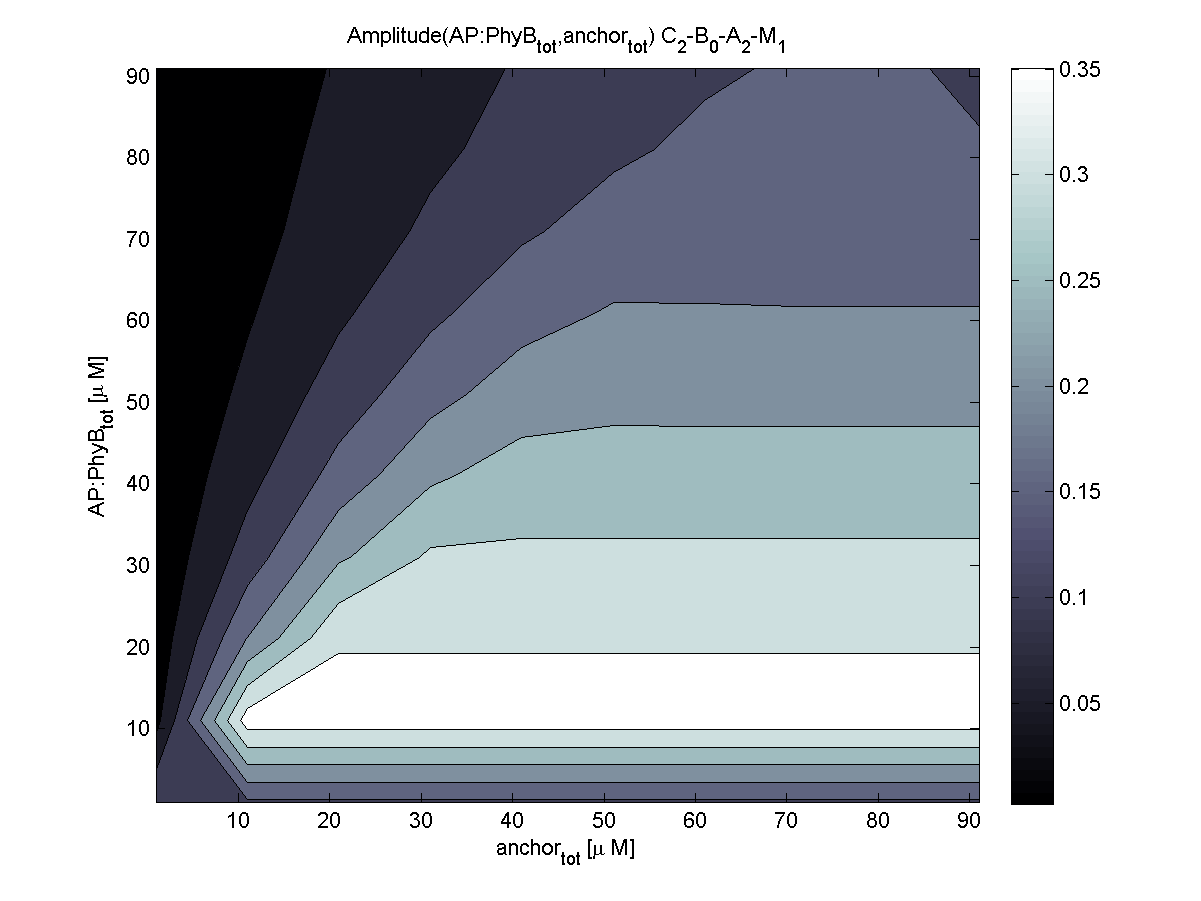
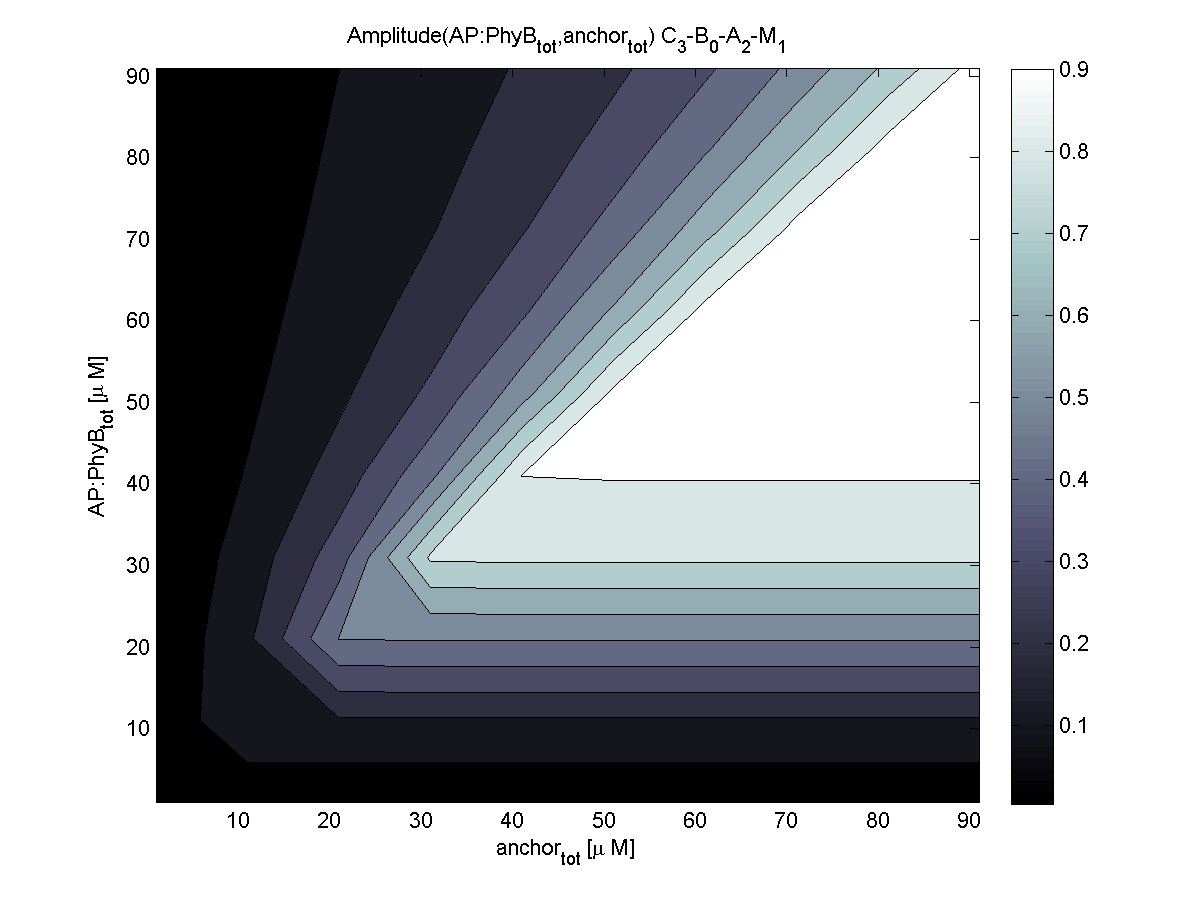
| - | |[[Image:ETHZ_Basel_models_evaluator_pif3.png|thumb|200px|''' | + | |[[Image:ETHZ_Basel_models_evaluator_pif3.png|thumb|200px|'''LSP? = PIF3.''' Coupling to PIF3 results in a much more sensitive area of [AP] vs. [anchor].]] |
|} | |} | ||
| + | |||
| + | PhyB is according to the evaluation the more robust choice compared to PIF3. Relative amplitude of both combinations is very similar. For this reasons, coupling to PhyB is first choice. | ||
=== 3. What is a good aspartate concentration? === | === 3. What is a good aspartate concentration? === | ||
Revision as of 09:37, 27 September 2010
Evaluation for wet laboratory
Goals of the evaluation
To determine the best possible parts for the biological implementation of E. lemming, the combined molecular model (Light switch - Chemotaxis) has been used to answer specific questions:
- Which receptor (Che) protein should be attacked?
- To which light-sensitive protein (PhyB, PIF3) should the receptor protein be linked?
- What is a good aspartate concentration?
- In what quantity should the anchor and binding light-sensitive protein be chosen?
Answers to this questions were able to decrease the experimental expenses significantly.
Evaluator
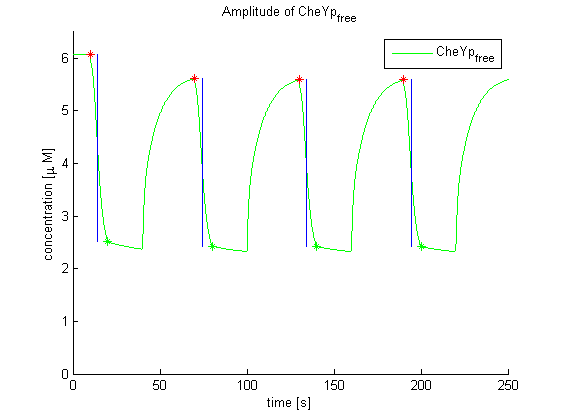
Evaluation of the results is a general optimization problem. The output variable in the case of E. lemming is the concentration of free and phosphorylated CheYp, because it is directly linked to the movement bias. To achieve a reasonable evaluation variable, the relative amplitude of free and phosphorylated CheYp was chosen to be maximized.
Because of the limited amount of different combination possibilities, it was chosen to evaluate all combinations. Parameter space of variable experimental implementation possibilities is shown in Table 1.
| Che? | LSP? | LSP?' | [Asp] | [AP] | [anchor] |
|---|---|---|---|---|---|
| CheR | PhyB | PIF3 | 0 uM | 0 uM | 0 uM |
| CheB | PIF3 | PhyB | 10^-6 uM | 25 uM | 25 uM |
| CheY | 10^-3 uM | 50 uM | 50 uM | ||
| CheZ | 75 uM | 75 uM | |||
| 100 uM | 100 uM |
The questions were answered in a hierarchical order, e.g. try Che1 before Che2.
Results
1. Which receptor (Che) protein should be attacked?
Although CheZ showed the highest relative amplitude, it was not chosen to be the first candidate, since it is not strictly located at the cell membrane and therefor would possibly not be suitable for our method of deactivation by changing local amount of active protein by spatial dislocation. The second best candidate in terms of relative amplitude was CheY, which was chosen to be the first target. CheR and CheB would have been possible backup targets in this order.
In addition, CheB and CheZ showed inverse activation / inactivation behavior than CheR and CheY upon light pulse induction. This means, that the activity would increase and therefore, this would have to be corrected by increasing the concentration of the proteins far above wild-type level to achieve an inverse effect of activation.
2. To which light-sensitive protein (PhyB, PIF3) should the receptor protein be linked?
PhyB is according to the evaluation the more robust choice compared to PIF3. Relative amplitude of both combinations is very similar. For this reasons, coupling to PhyB is first choice.
3. What is a good aspartate concentration?
4. In what quantity should the anchor and binding light-sensitive protein be chosen?
Evaluation for information processing
Goal of the evaluation
Main goal of the evaluation for information processing was to determine time constants of E. lemming by using the combined molecular model (Light switch - Chemotaxis).
 "
"