Team:ETHZ Basel/Modeling/Imaging
From 2010.igem.org
Setup of the Microscope
A fluorescence microscope with motorized x, y and z control, a motorized shutter and a 60× lens is used with appropriate fluorescence filters for the fluorescence signals. Light-emitting diode arrays are installed as light sources for red light (660 nm) and far-red light (748 nm) pulses.
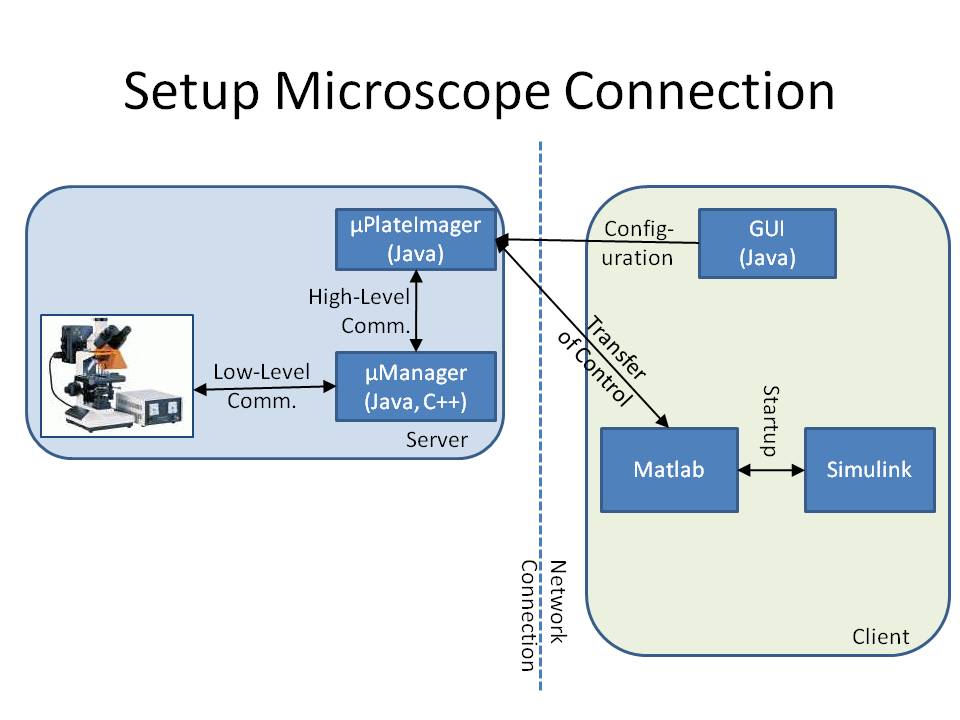
Control of the Microscope
The microscope is connected to a workstation using the core drivers and interfaces of μManager (see Stuurman et al. (2007) or [1]). To provide a mechanism to change the cell's input signal depending on its fluorescence signal, we developed the microscope software μPlateImager, which enables for parallel acquisition of images and the modification of light input signals. μPlateImager uses the Java interface of the μManager core to control the microscope and can be configured by a separate platform-independent visual user interface. μPlateImager uses the undocumented Java MATLAB Interface (JMI) to connect to Matlab (The MathWorks, Natick, MA) based on the open source project matlabcontrol (see [2]). The microscope thus can be closely controlled by standard Matlab scripts.
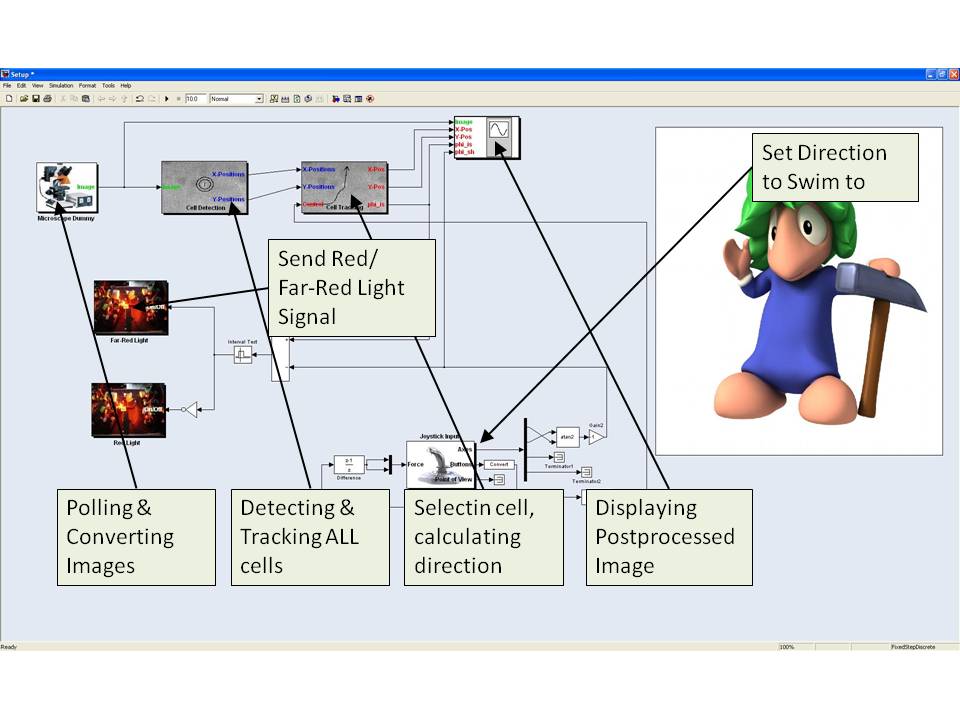
Image Analysis
The direction of the cell is automatically compared to the direction it should go. This direction can be intuitively defined by the user using a joystick. The force feedback functionality of the joystick is used to give the user an intuitive feedback of the current direction of the E. lemming. If the difference between the actual direction of the E. lemming and the direction the user defined is too high, tumbling is automatically induced by a red light (660nm) pulse. Otherwise tumbling is supressed by a far red light (748nm). Alternatively the user can induce the pulses directly using the buttons of the joystick.
If the cell is moving out of the image the microscope moves automatically such that the cell is always approximately in the center of the image.
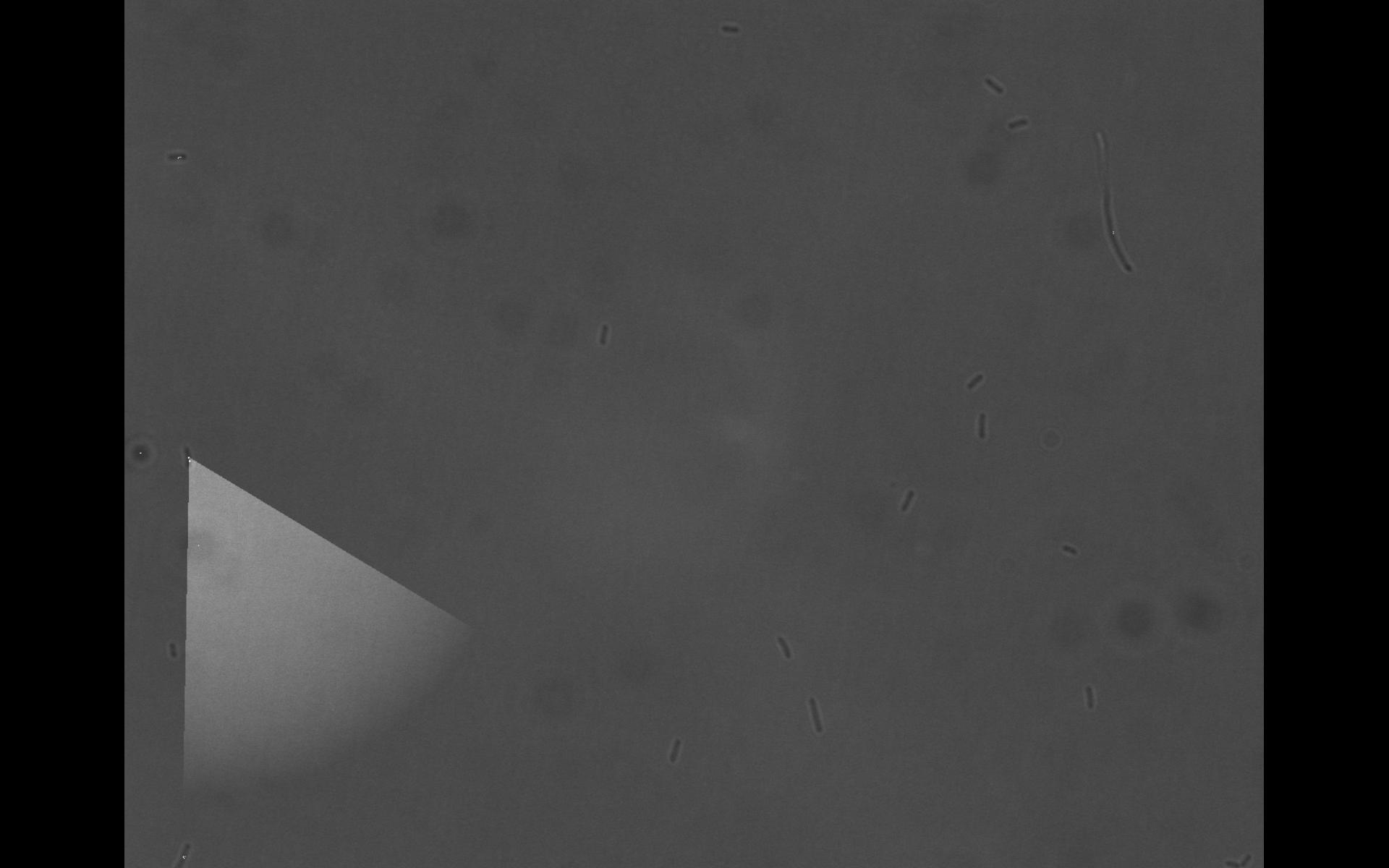
First Imaging
To test the setup of the microscope we imaged a ΔCheR strain. The videos of the imaging can be found below. Please remark that the quality of the movie was decreased and that the setup will be enhanced so that the final movies will look nicer (yet just a quick & dirty trial).
The cells were placed in a 50 μm (?) high flow chamber (details on the setup later). The movies were made out of bright field images. The excitation was 60ms, the period something around 1/7s, pretty much the fps rate of the movie (we can go heigher, but yet we already produced like 2 GB of raw images for every movie). Every image has the size of 1344 x 1024 pixels, 12 or 16 bit grayscale (I forgot what I have chosen, probably we will reduce it anyway later to speed up the imaging pipeline). For both movies the images were made out-of-focus to simplify cell detection.
 "
"