Team:ETHZ Basel/Modeling
From 2010.igem.org
(→Mathematical Modeling Overview) |
(→Mathematical Modeling Overview) |
||
| Line 12: | Line 12: | ||
== Model implementation == | == Model implementation == | ||
=== Submodels === | === Submodels === | ||
| - | + | First of all submodels for the individual processes of E. lemming have been identified by literature research, implemented, corrected and adapted to our needs. Where we could not rely on established models, we started modeling on our own and calibrating the model with regard to literature knowledge. | |
| - | |||
* [[Team:ETHZ_Basel/Modeling/Light_Switch|'''Light Switch''']]: a deterministic molecular model based on the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3. | * [[Team:ETHZ_Basel/Modeling/Light_Switch|'''Light Switch''']]: a deterministic molecular model based on the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3. | ||
* [[Team:ETHZ_Basel/Modeling/Chemotaxis|'''Chemotaxis Pathway''']]: two deterministic molecular models of the chemotaxis pathway. | * [[Team:ETHZ_Basel/Modeling/Chemotaxis|'''Chemotaxis Pathway''']]: two deterministic molecular models of the chemotaxis pathway. | ||
| Line 21: | Line 20: | ||
=== Merged models === | === Merged models === | ||
[[Image:ETHZ_Basel_models_overview_comb.png|thumb|400px|'''Combined models.''' Coupled individual models for the simulation of the entire process and their interfaces.]] | [[Image:ETHZ_Basel_models_overview_comb.png|thumb|400px|'''Combined models.''' Coupled individual models for the simulation of the entire process and their interfaces.]] | ||
| - | + | Secondly we combined the submodels stepwise to more comprehensive models that we could use to address different important questions to: | |
| - | + | * [[Team:ETHZ_Basel/Modeling/Combined#Light_switch_-_Chemotaxis |'''Light switch - Chemotaxis''']]: used for design of experiment for the biology lab and the information processing. | |
| - | + | ||
| - | * [[Team:ETHZ_Basel/Modeling/Combined#Light_switch_-_Chemotaxis |'''Light switch - Chemotaxis''']]: used | + | |
* [[Team:ETHZ_Basel/Modeling/Combined#Light_switch_-_Chemotaxis_-_Movement |'''Light switch - Chemotaxis - Movement''']]: complete model of E. lemming. | * [[Team:ETHZ_Basel/Modeling/Combined#Light_switch_-_Chemotaxis_-_Movement |'''Light switch - Chemotaxis - Movement''']]: complete model of E. lemming. | ||
Revision as of 12:34, 26 October 2010
Mathematical Modeling Overview
A complex mathematical model of E. lemming from both literature inspired and self developed submodels was created. The final implementation combines deterministic molecular models of the chemotaxis pathway, the light switch and a probabilistic model for the bacterial movement in one model.
This model was used to reduce wet laboratory experiments by identify molecular targets and to create a simulative test bench for the information processing algorithms.
Model implementation
Submodels
First of all submodels for the individual processes of E. lemming have been identified by literature research, implemented, corrected and adapted to our needs. Where we could not rely on established models, we started modeling on our own and calibrating the model with regard to literature knowledge.
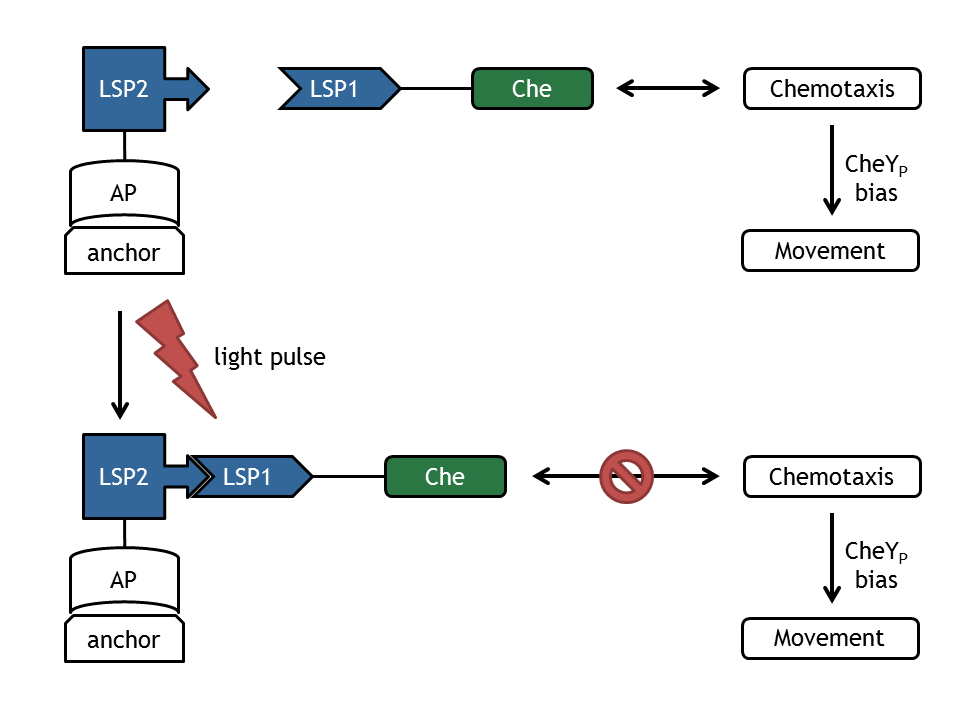
- Light Switch: a deterministic molecular model based on the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3.
- Chemotaxis Pathway: two deterministic molecular models of the chemotaxis pathway.
- Bacterial Movement: a self developed stochastic model of E. coli movement on basis of the CheYp bias.
Merged models
Secondly we combined the submodels stepwise to more comprehensive models that we could use to address different important questions to:
- Light switch - Chemotaxis: used for design of experiment for the biology lab and the information processing.
- Light switch - Chemotaxis - Movement: complete model of E. lemming.
Experimental Design
Insights for wet laboratory
Since the design of E.lemming implies the existence of many possible biological combinations of the fundamental parts, the first steps in the modeling of our system were directed towards reducing the actual number of combinations implemented in the wetlab. Furthermore, by using the combined molecular models for in silico evaluation of the best possible devices, we derived theoretical results on choosing the biological parts which maximize the chance of a functional final ensemble.
Wet laboratory evaluation results have showed that molecular modeling and experimental biology can interwork to gain new insight on both aspects of our project.
Insights for information processing
In order to adjust the controller to have optimal light pulse rates, the combined molecular model has been used to determine the corresponding time constants.
Information processing evaluation results provide further information on how this task has been accomplished.
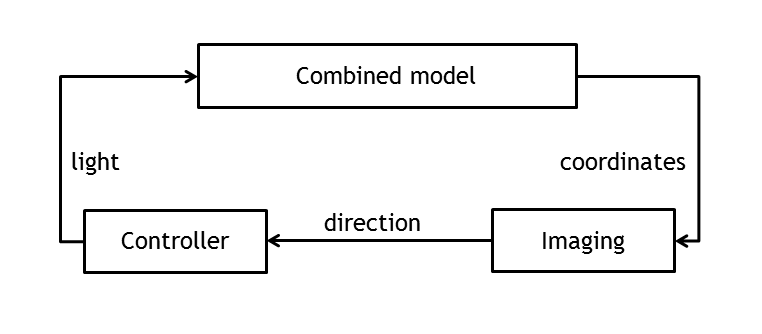
Test bench for information processing
In order to create a first test bench for the information processing pipeline, the combined model has been used to set up and evaluate the controller.
By providing input ports for the actual and the desired movement direction of the bacterium and boolean output ports for both light pulses (red light/far red light), it was possible to time - dependently control the E. lemming and close the loop in simulating the overall network.
 "
"