Team:ETHZ Basel/InformationProcessing/InformationFlow
From 2010.igem.org
Information Flow
Information Acquisition
μPlateImager
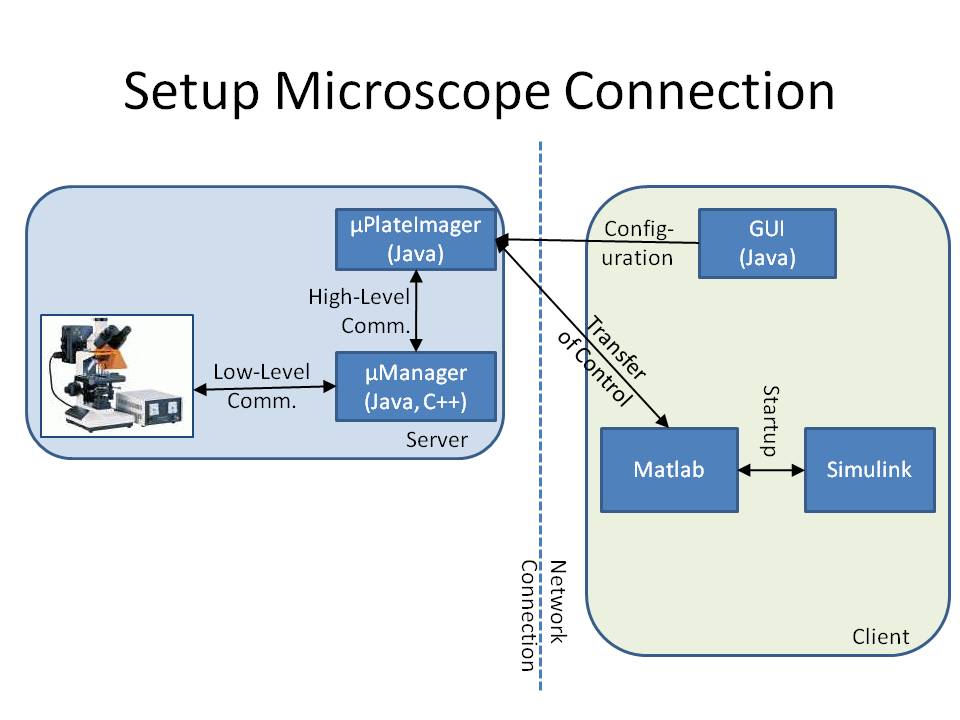
The microscope is connected to a workstation using the core drivers and interfaces of μManager (see [1] or [2]). To provide a mechanism to change the cell's input signal depending on its fluorescence signal, we developed the microscope software μPlateImager, which enables the parallel acquisition of images and the modification of light input signals.
μPlateImager uses the Java interface of the μManager core to control the microscope and can be configured by a separate platform-independent visual user interface. Since the communication with the microscope already requires a significant amount of system resources, it was necessary to swap the image processing, cell detection, and controller part to a second workstation to increase the possible frame rate.
μPlateImager can be controlled over the network or the Internet by a GUI (graphical user interface). This GUI uses the yet undocumented Java MATLAB Interface (JMI) to start up a Matlab process, based on the open source project matlabcontrol (see [3]). It furthermore starts up a Simulink model and transfers the microscope control to several of the blocks of the Lemming Toolbox.
Information Processing
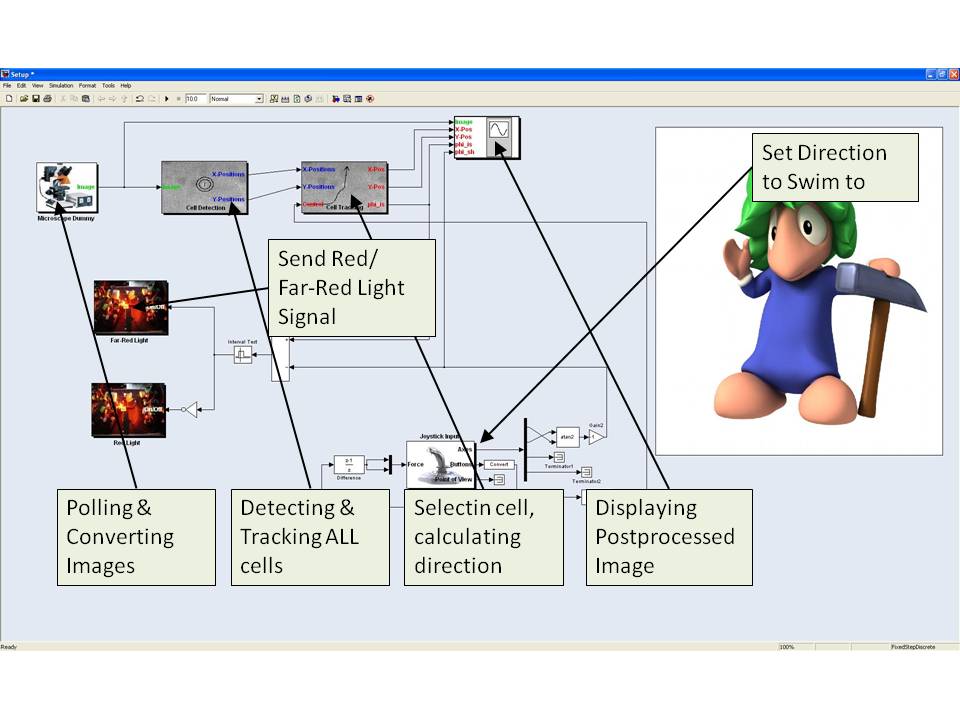
First, the microscope block triggers μPlateImager and thus indirectly the microscope to make an out-of focus image and sends this image to the cell detection block. Based on a user definable threshold the cell detection block returns the positions of the cells having the highest score. These positions are send to the Cell Tracking block, which compares the positions of the detected cells with those of in previous frames detected cells. If the distance of a currently detected cell from its position in one of the previous frames is smaller than a threshold, the block assigns the same unique ID (UID) to the cell, otherwise a new UID is generated. Furthermore the cell tracking algorithm allows the cell to persist for several successive frames if it gets lost. The cell positions together with their UIDs is send to the Cell Selection block. This block determines the direction of the currently selected cell by calculating its change of its actual position compared to its position a few frames before. Furthermore it allows the user to switch between the cells with the help of the buttons of a connected joystick.
With the joystick the user furthermore defines a reference direction, which serves together with the actual direction of the E. lemming as the input for the Controller block. This controller calculates, depending on the actual and previous direction of the E. lemming and the reference direction if red light or far-red light should be activated. This information is passed to the Red Light and Far-Red Light blocks, which trigger μManager and therewith the microscope to activate or deactivate the light sources.
The Stage Control block gets as input the current position of the E. lemming and decides if the E. lemming is on the verge of moving out of the field of view. If so, it induces the x/y-stage of the microscope to change for a defined amount of μm. The current stage position is passed to the Cell Tracking Block as an additional input to allow for the tracking of cells over a long distance.
Finally, several of the previous described blocks outputs are passed to the Visualization block, which displays all information on the computer screen or the beamer.
References
[1] Stuurman et al. 2007
 "
"