Team:ETHZ Basel/Biology/Cloning
From 2010.igem.org
Cloning strategy for the construction of our Biobricks
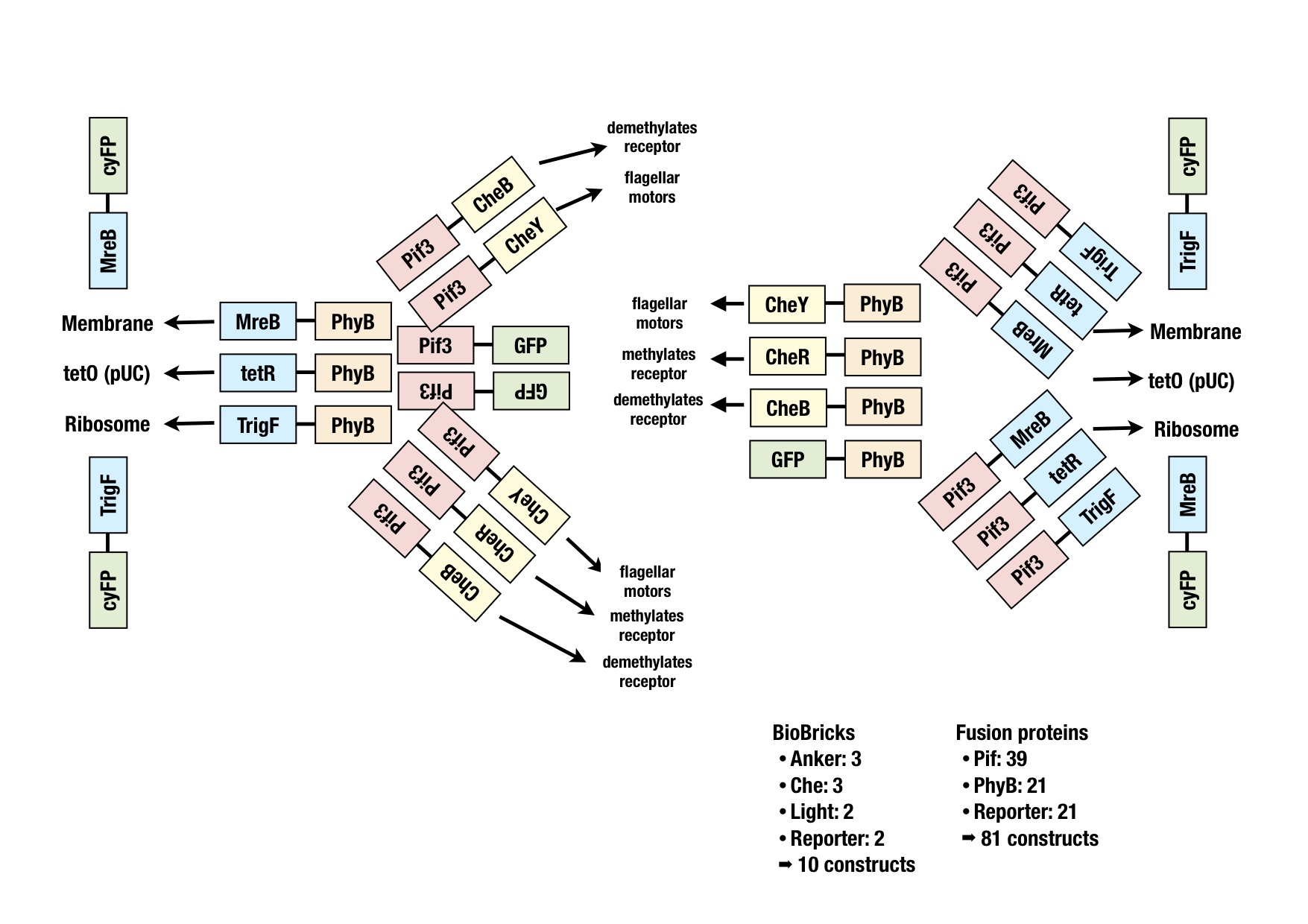
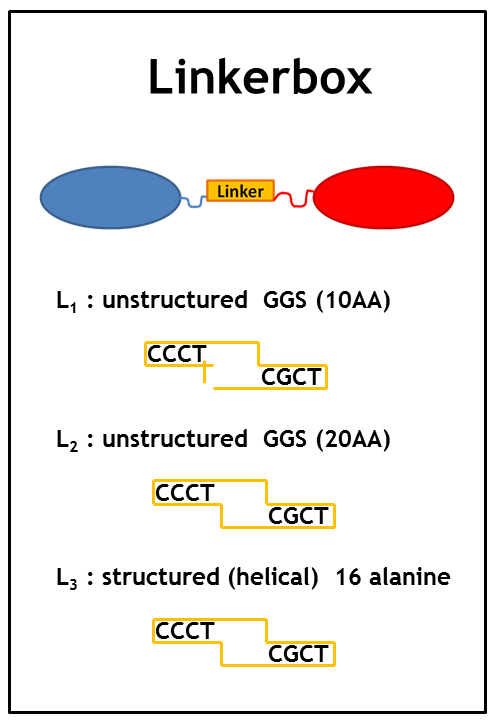
When starting the project it was hard to know which implementation strategy would work the best: Which type of anker would work the best? Which Che protein should be chosen and should it be C-terminally or N-terminally linked to the LSP? Could even the type of linker make a difference?
Therefore we planned to generate several fusion proteins with different linkers to start a model base experimental design in parallel and to find out the best working system in the end. The picture graphically represents all the fusion proteins we have now in our wetlab production pipeline. For efficient cloning of such a huge amount of fusion proteins we decided to apply the cloning strategy BBF RFC28: A method for combinatorial multi-part assembly based on the Type IIs restriction enzyme AarI [1]. The advantage of this strategy is that we can simultaneously clone up to 3 different inserts into one single expression vector. In the following section we give an overview of how this was achieved.
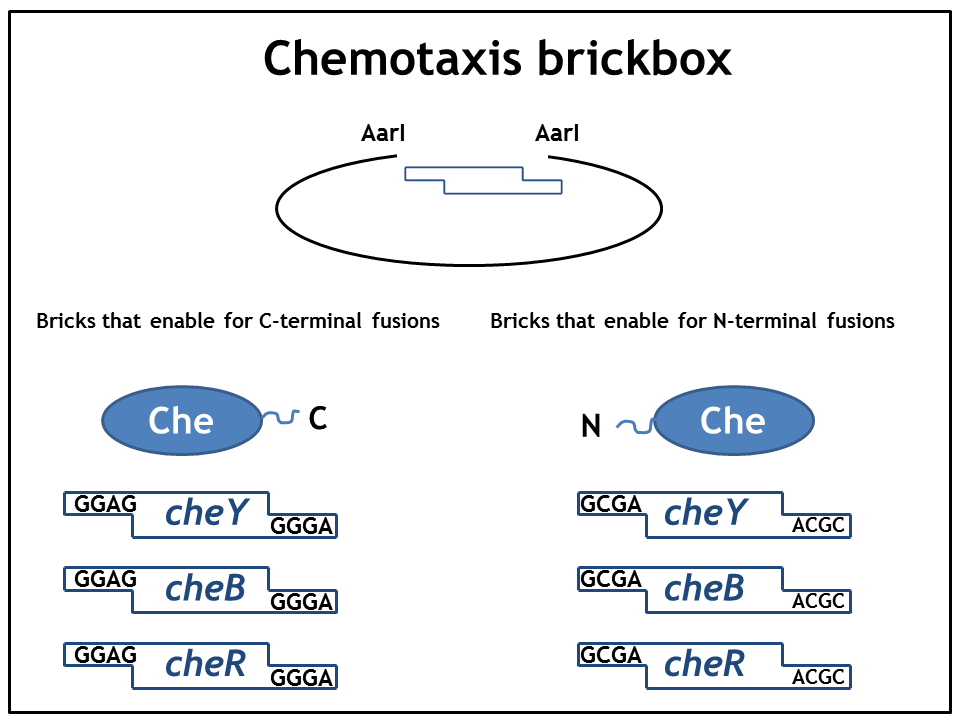
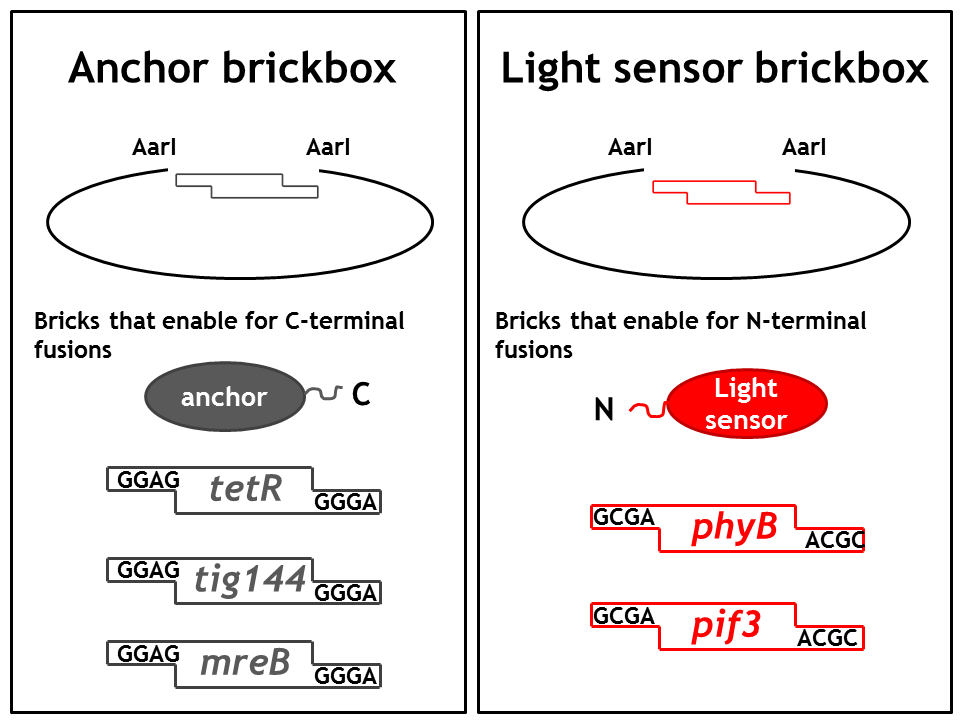
1. Step: Construction of brickboxes which enable the generation of fusion proteins
Those Parts were generated by PCR. We used primers, (specified in the BBF RFC28 manual) which were subcloned into the storage vector pSEVA132 (Victor de Lorenzo's lab, KanR, pBBR1 ori) by blunt end ligation. The plasmid pSEVA132 enables us to pre-screen the transformed colonies by blue white screening. This boosted the production of the brickbox parts, as we could pick the "right" colonies for the subsequent mini-prep procedure! Finally, the generated parts were verified by AarI digest and sequencing. Due to the presence of rare codons in the sequence of PhyB and Pif3, these two genes had to be codon-optimized (ordered from GeneArt). As the succesful implementation of our E. Lemming's pathway mainly relies on two protein fusions (the "anchor-light sensor" and "light sensor-che protein" fusion), we constructed two expression vectors (we call them working vectors), which enabled us to simultaneously express the two fusion proteins in the same plasmid.
- Working vector 1 is a derivative of pSEVA132 conferring resistance to ampicillin and replicating with ori pBB1.
- Working vector 2 is a derivative of pSEVA421 expressing a spectinomycin resistance cassette and replicating with ori RK2. The gene for the repressor AraC and the corresponding ParaBAD promotor/operator sites were introduced into both vectors followed by an insert, which is flanked by AarI-recognition sites. Digest with AarI releases this insert and generates a vector with assembly-compatible overhangs.
2. Step: Assembly of fusion proteins using the generated brickboxes
The following image illustrates the assembly of a fusion protein. In the section "Implementation" the constructed fusion proteins, required for the implementation of the synthetic network, are described. But how should the fusion process look like? Should we use the C-term or the A-term? Would this have an effect on the functionality of the biobricks? The decision which fusion strategy to choose, could be taken thanks to some models. The modeling group successfully tackled this predominant question and achieved to "in silico" formulate the crucial system parameters required for a working system.
Linking BBF RFC28 to the Tom Knight's original assembly standard (OAS) [2]
General scheme for the design and construction of Tom Knight's OAS-compatible fusion proteins
BBF RFC28 is a method for combinatorial multi-part assembly based on the Type II restriction enzyme AarI and it is a highly efficient method for constructing fusion proteins. As we have a great number of constructs to generate, efficiency in the cloning process is vital for the project. Unfortunately it is not compatible with the general iGEM standardized biobricks. As a consequence, the fusion proteins are not compatible with Tom Knight's original assembly standard as they don't contain the required prefix and suffix sequences.
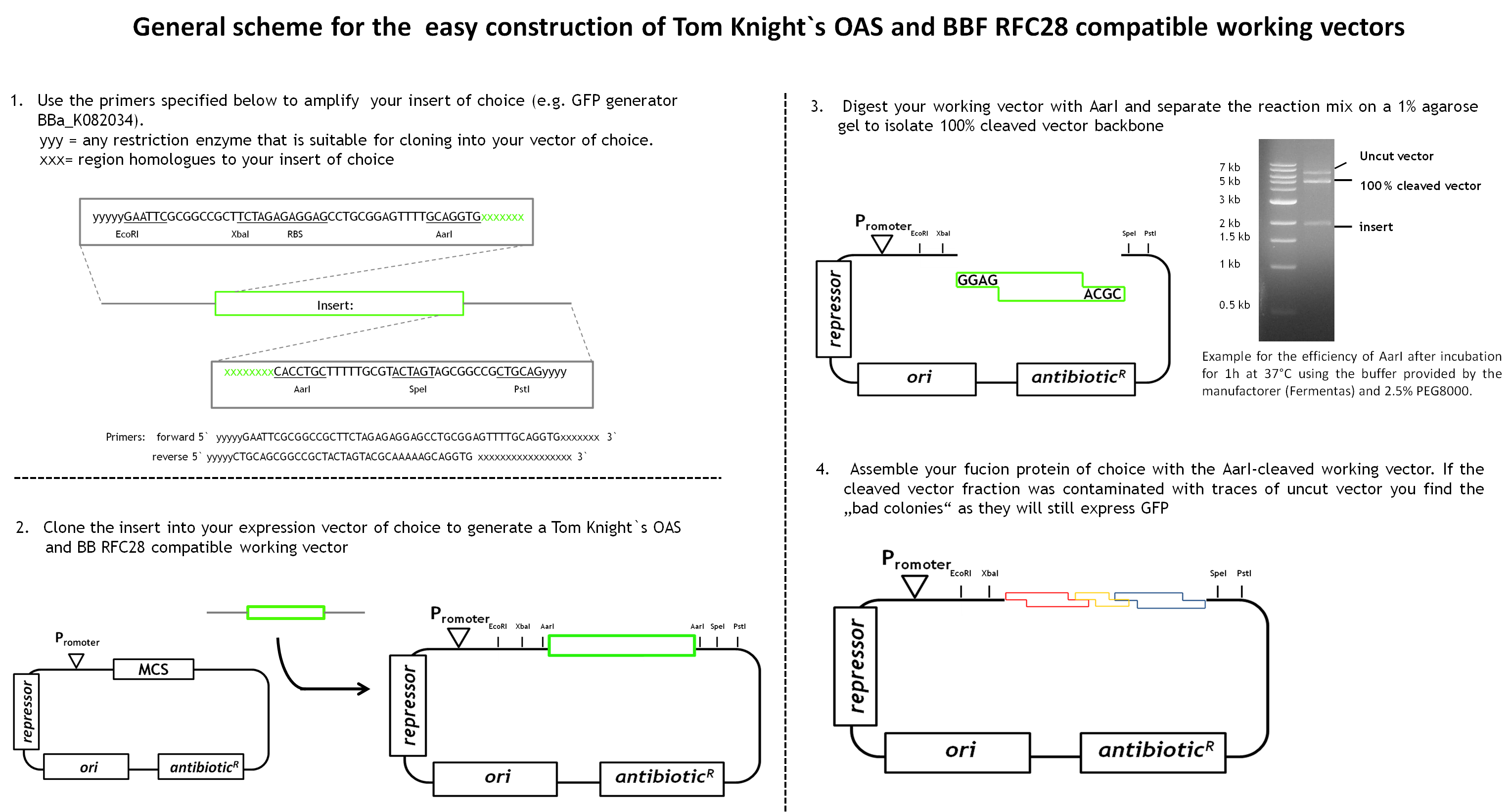
During our project we learned a lot about the cloning of fusion proteins using the BBF RFC28 assembly method-especially how to make it work efficiently. We would like to share our experience with you, especially as we worked out a general design method for the facilitated construction of Tom Knight's OAS and BBF RFC28 compatible working vectors (the expression vector that is finally used for the assembly) that can be used for the final assembly step. Our vector design includes a GFP-mediated screening system for screening clones containing the fusion protein.
As we had already started the cloning procedure before learning about the little difficulties of the assembly method and before making up our minds on how to improve it and link it to the Tom Knight's BBF, our fusion proteins were not cloned into a Tom Knight's OAS-compatible working vector. Nevertheless, the design scheme that is outlined below is under construction.
In general the working vector needs to contain an insert that is flanked by AarI-sites. Those cleavage sites must to be designed such that AarI-digestion releases the insert and generates a vector backbone with 5` overhangs compatible with the BBF RFC28 standard. AarI does not work with the same high efficiency on every cleavage site. Therefore, the digest needs to be separated on a 1% agarose agarose gel to isolate 100% cleaved vector backbone from uncut vector proportions and the insert. By choosing a constitutively expressed reporter protein as insert, positive clones containing the assembled protein can be distinguished from clones containing the original insert (traces of uncut vector might still be present in the cleaved vector fraction) by green/white screening. Constitutive expression of the reporter is very convenient as adding an inducer to the selection plates is no longer necessary. This gives total freedom of choice for the fusion protein expression system.
To contribute to the improvement of the registry we screened it for a reporter that might full fill these requirements and was not yet characterized. We found the GFP generator BBa_K082034 as potential target and characterized it therefore. You find the results below.
References
[1] [http://dspace.mit.edu/handle/1721.1/46721 BBF RFC 28: A method for combinatorial multi-part assembly based on the Type IIs restriction enzyme AarI]
[2] [http://dspace.mit.edu/handle/1721.1/21168 Idempotent Vector Design for Standard Assembly of Biobricks]
 "
"