Team:ETHZ Basel/Biology/Archeal Light Receptor
From 2010.igem.org
Archeal Light Receptor
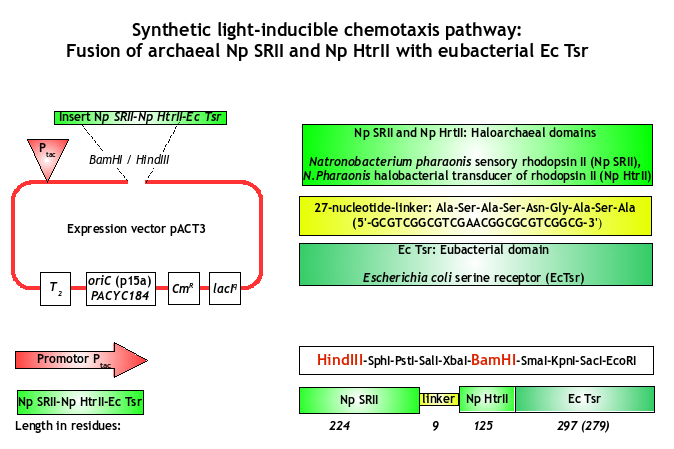
Parallely to the light-sensitive Pif3/PhyB-system, another implementation strategy caught our attention: The generation of our E. lemming by the fusion of an archean photoreceptor to a bacterial chemotactic transducer. This was successfully demonstrated by Jung et al. in 2001 [1], who fused the Natronobacterium pharaonis NpSRII (Np seven-transmembrane retinylidene photoreceptor sensory rhodopsins II) and their cognate transducer HtrII to the cytoplasmic domain of the chemotaxis transducer EcTsr of Escherichia coli.
Rhodopsins are photoreactive, membrane-embedded proteins, which are found not only in archaea, but in eubacteria and microbes as well. In Natronobacterium pharaonis, the NpSRII contains a domain of seven membrane-spanning helices, which carry out two distinct functions: Firstly, they serve as photo-inducible ion-pumps and secondly, as actors in the chemotaxis signaling network. This was proposed by Spudich et al. in 2001.
Encouraged by this work, we cloned the archeal photoreceptor NpSRII as biobrick into the standardized vector pSB1C3 BBa_K422001. For the expression (and experimental purposes), we used pACT3 (BamHI/HindIII) as plasmid backbone, which has a CmR gene (Chloramphenicol resistance) and is IPTG-inducibile (Isopropyl-ß-D-thiogalactopyranosid).
Primers for pACT3
Name: AR-BamHI-forward Scale: 0.04 umol Purification: Desalted 5' Mod.: None Inner Mod.: None 3' Mod.: None Type: DNA Sequence (5'-3'): GTG GAT CCA AGG AGA TAT ACA TAT GGT TGG TCT GAC CAC CCT G Length: 43 Aliquots: NONE
Name: AR-Hind-reverse
Scale: Genomics
Purification: Desalted
5' Mod.: None
Inner Mod.: None
3' Mod.: None
Type: DNA
Sequence (5'-3'): GCA AGC TTT TAA CCG CTA TAA ATT G
Length: 25
Aliquots: NONE
Sequence
LOCUS 1046531_aercheal_taxisphusio 1964 bp DNA
FEATURES Location/Qualifiers
CDS 27..1937
/label="aercheal_taxisphusio"
ORIGIN
GAGCTCGAATTCGCGGCCGCTTCTAGATGGTTGGTCTGACCACCCTGTTTTGGCTGGGTG
CAATTGGTATGCTGGTTGGCACCCTGGCATTTGCATGGGCAGGTCGTGATGCAGGTAGCG
GTGAACGTCGTTATTATGTTACCCTGGTTGGTATTAGCGGTATTGCAGCAGTTGCATATG
CAGTTATGGCACTGGGTGTTGGTTGGGTTCCGGTTGCAGAACGTACCGTTTTTGTTCCGC
GTATTGATTGGATTCTGACAACTCCGCTGATTGTGTACTTCCTGGGTCTGCTGGCAGGTC
TGGATAGCCGTGAATTTGGTATTGTTATTACCCTGAATACCGTTGTTATGCTGGCAGGTT
TTGCCGGTGCAATGGTTCCGGGTATTGAACGTTATGCACTGTTTGGTATGGGTGCAGTTG
CATTTATTGGCCTGGTTTATTATCTGGTTGGTCCGATGACCGAAAGCGCAAGCCAGCGTA
GCAGCGGTATTAAAAGCCTGTATGTTCGTCTGCGTAATCTGACCGTTGTTCTGTGGGCAA
TTTATCCGTTTATTTGGCTGCTGGGTCCGCCTGGTGTTGCACTGCTGACCCCGACCGTTG
ATGTTGCACTGATTGTTTATCTGGATCTGGTTACCAAAGTGGGCTTTGGTTTTATTGCAC
TGGATGCAGCAGCGACCCTGCGTGCAGAACATGGTGCAAGCGCAAGCAATGGTGCCAGCG
CCAGCCTGAATGTTAGCCGTCTGCTGCTGCCTGGTCGTGTTCGTCATAGCTATACCGGTA
AAATGGGTGCAACCTTTGCATTTGTTGGTGCACTGACCGTTCTGTTTGGTGCAATTGCAT
ATGGTGAAGTTACCGCAGCAGCAGCCACCGGTGATGCAGCAGCCGTTCAAGAAGCAGCAG
TTAGCGCAATTCTGGGTCTGATTATTCTGCTGGGTATTAATCTGGGTCTGGTTGCAGCCA
CCCTGGGTGGTGATACCGCAGCAAGCCTGAGCACCCTGGCAGCAAAAGCAAGCCGTATGG
GTGATGGTGATCTGGATGTTGAACTGGAAACCCGTCGTGAAGATGAAATTGGTGATCTGT
ATGCAGCCTTTGATGAACTGAAACGCATTAAAATTGTGACCAGCCTGCTGCTGGTTCTGG
CAGTTTTTGGTCTGCTGCAACTGACCAGCGGTGGTCTGTTTTTTAATGCACTGAAAAACG
ATAAAGAAAATTTTACCGTGCTGCAAACCATTCGCCAGCAGCAGAGCACCCTGAATGGTA
GCTGGGTTGCCCTGCTGCAAACCCGTAATACCCTGAATCGTGCAGGTATTCGTTATATGA
TGGACCAGAATAATATTGGTAGCGGTAGCACCGTTGCAGAACTGATGGAAAGCGCCAGCA
TTAGCCTGAAACAGGCAGAAAAAAACTGGGCAGATTATGAAGCACTGCCTCGTGATCCGC
GTCAGAGCACCGCAGCAGCCGCAGAAATTAAACGTAACTATGATATCTACCATAACGCAC
TGGCAGAACTGATTCAGCTGCTGGGTGCAGGTAAAATCAACGAATTTTTTGATCAGCCGA
CCCAGGGTTATCAGGATGGTTTTGAAAAACAGTATGTGGCCTACATGGAACAGAATGATC
GTCTGCATGATATTGCCGTGAGCGATAATAATGCAAGCTATAGCCAGGCAATGTGGATTC
TGGTTGGTGTTATGATTGTTGTTCTGGCCGTTATTTTTGCCGTGTGGTTTGGTATTAAAG
CAAGCCTGGTTGCACCGATGAATCGTCTGATTGATAGCATTCGTCATATTGCCGGTGGTG
ATCTGGTTAAACCGATTGAAGTTGATGGCAGCAATGAAATGGGTCAGCTGGCAGAAAGCC
TGCGTCATATGCAGGGTGAACTGATGCGTACCGTTGGTGATGTTCGTAATGGTGCAAATG
CAATTTATAGCGGTTAATACTAGTAGCGGCCGCTGCAGGGTACC
//
Chrystal structure of NpSRII
Np SRII
Model of the quaternary Np HtrII/Np SRII complex
Sensory Rhodopsin II photocycle
References
[2] Hartmut Luecke1 and Brigitte Schobert, Janos K. Lanyi, Elena N. Spudich, John L. Spudich. Crystal Structure of Sensory Rhodopsin II at 2.4 Angstroms: Insights into Color Tuning and Transducer Interaction. Science Express. 2001; 293;5534
[3] [http://www.ingentaconnect.com/content/els/00145793/1997/00000420/00000001/art01487: Shimono K, Iwamoto M, Sumi M, Kamo N.: Functional expression of pharaonis photorhodopsin in Escherichia coli. FEBS Lett 1997; 22]
[6] Jones, P. C., and R. H. Fillingame. 1998. Genetic fusions of subunit c in the F0 sector of H+-transporting ATP synthase. Functional dimers and trimers and determination of stoichiometry by cross-linking analysis. J. Biol. Chem. 273:29701-29705
 "
"