Team:ETHZ Basel/Biology
From 2010.igem.org
(→generation of storage vectors) |
|||
| Line 1: | Line 1: | ||
| - | |||
== Overview == | == Overview == | ||
| Line 104: | Line 103: | ||
=== generation of ''E. coli'' mutants === | === generation of ''E. coli'' mutants === | ||
| + | |||
| + | == experimental plan == | ||
Revision as of 09:25, 29 July 2010
Contents |
Overview
The goal of our iGEM project E.lemming is to take over the control of the tumbling frequency of E. coli. This is achieved by reversibly localizing certain elements of the chemotactic pathway (che-proteins) and thus affecting their activity.
For localization we use the PhyB-PIF3 system found in plants. PhyB or PIF3 is fused to a che-protein while the other one is fused to a localized protein (localizer) within the cell. Upon a light stimulus of a certain wavelength PhyB is activated, binds PIF3 and thereby localizes the che-protein. A light stimulus of a different wavelength reverses this process.
In order to increase the probability that the system will work we will try out a lot of combinations of several proteins:
- che-proteins: cheY, cheB and cheR
- localizer: trigger factor (binds the ribosome), MreB (actin analogue), tetR (binds tetO)
The goal of the wet lab team is to implement this localization system into E. coli.
Cloning strategy
As we plan to generate several fusion proteins with different linkers, we decided to use the cloning strategy BBF RFC 28: A method for combinatorial multi-part assembly based on the Type IIs restriction enzyme AarI (http://dspace.mit.edu/handle/1721.1/46721). The advantage of this strategy is that we can clone up to 3 different inserts into one vector simultaneously in a 96 well format.
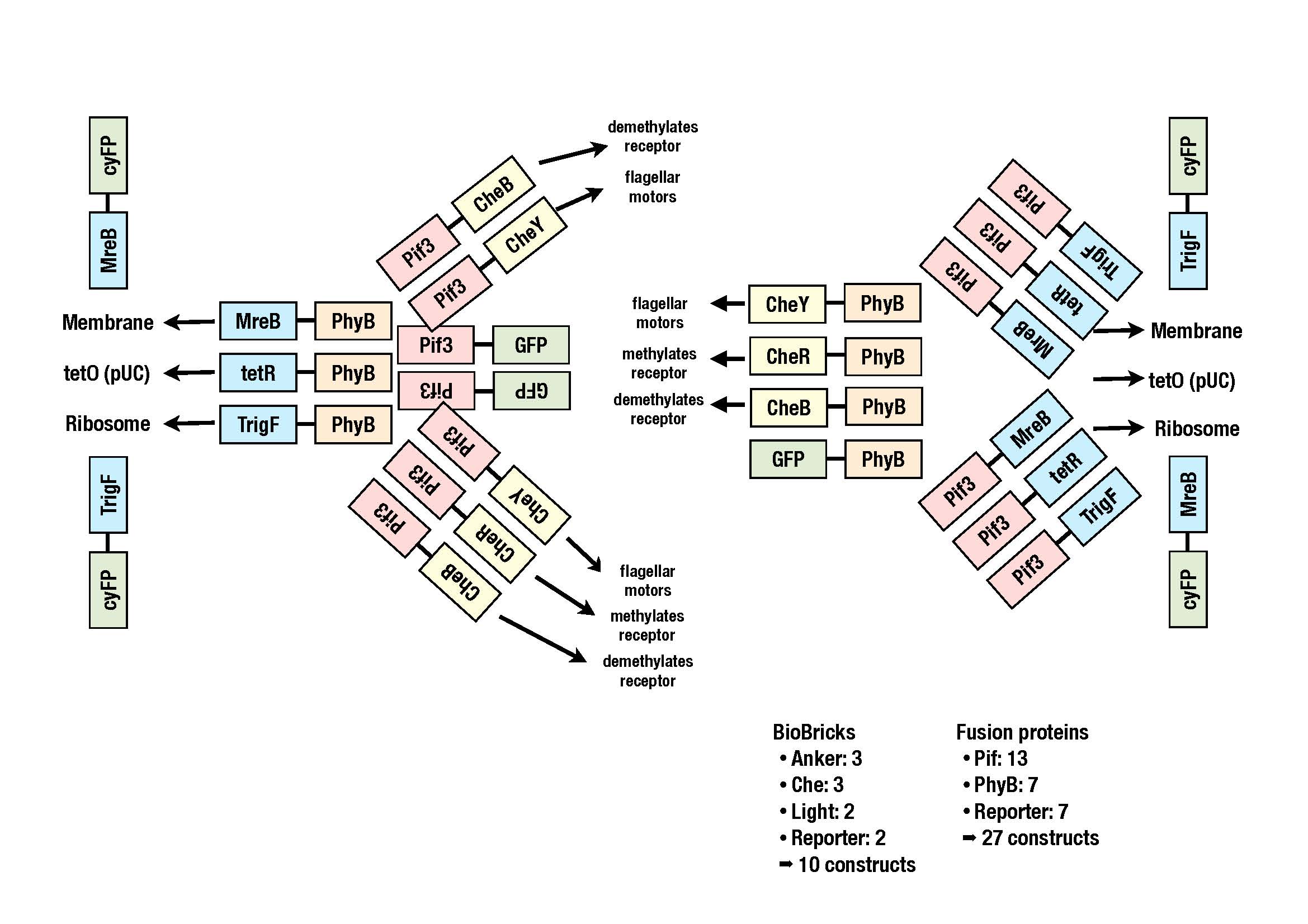
Parts
Currently we are working on putting all the BioBricks into a storage vector. The image shows all the constructs we plan to clone.
working process
generation of parts
generation of storage vectors
We use the vector pSEVA of Victor de Lorenzo's lab. It has a kanamycin resistance and a BBR1 origin.
The working process for the generation of the storage vectors is as follows:
Ordering of primers (if template is available) -> PCR -> clean-up of PCR product -> ligation into storage vector -> transformation of competent cells -> plating of cells -> selection of clones (blue-white-screening) -> sequencing
For proteins for which no template is available we let the let the genes synthesize directly.
| Protein | terminus on fusion | status |
|---|---|---|
| CheY | N | plating of cells |
| CheY | C | plating of cells |
| CheR | N | plating of cells |
| CheR | C | plating of cells |
| CheB | N | plating of cells |
| CheB | C | plating of cells |
| TrigF | N (binds N-terminal to ribosome) | primers recieved |
| MreB | N | primers ordered |
| MreB | C | primers ordered |
| tetR | N | primers recieved |
| tetR | C | primers recieved |
| PhyB | N | primers ordered |
| PhyB | C | primers ordered |
| PhyB | N | primers ordered |
| PhyB | C | primers ordered |
| PIF3 | N | primers ordered |
| PIF3 | C | primers ordered |
| cyFP | N | |
| cyFP | C | |
| GFP | N | |
| GFP | C |
 "
"