Team:Cambridge/Tools/Gibson
From 2010.igem.org
(Difference between revisions)
| Line 12: | Line 12: | ||
{{:Team:Cambridge/Templates/rightpic|src=Gibthon-dc-beta.png}}<html> | {{:Team:Cambridge/Templates/rightpic|src=Gibthon-dc-beta.png}}<html> | ||
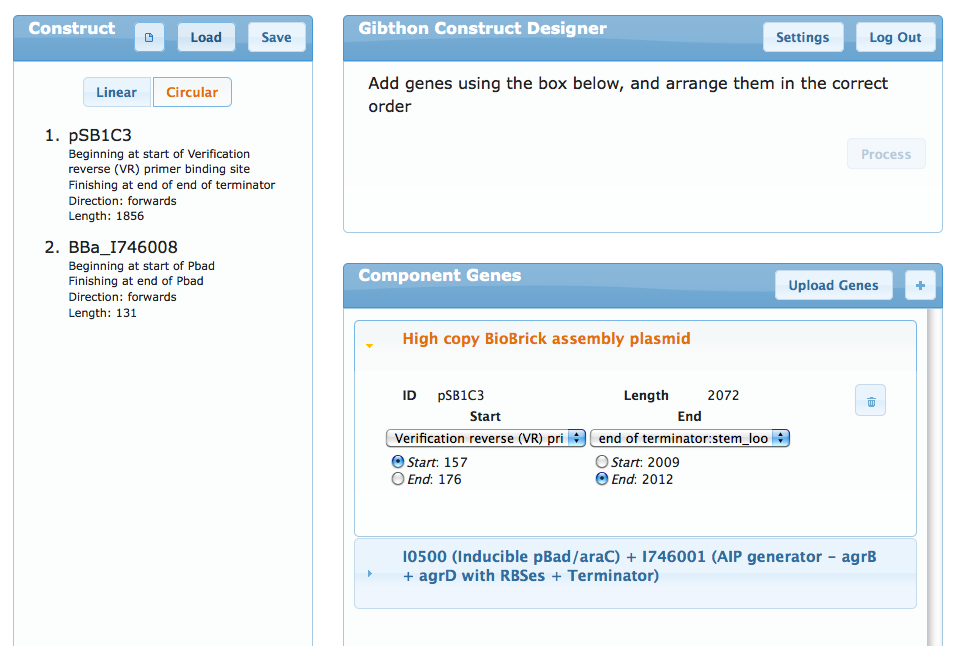
Gibthon's older brother, and much more powerful. With this, you can design your construct from its component genes and automatically generate the optimal primers. The tool has access to the Parts Registry Database, the NCBI's Nucleotide database, as well as accepting genbank formatted files from the user. The user is able to store constructs and their own genes for recovery later.<br /><br /> | Gibthon's older brother, and much more powerful. With this, you can design your construct from its component genes and automatically generate the optimal primers. The tool has access to the Parts Registry Database, the NCBI's Nucleotide database, as well as accepting genbank formatted files from the user. The user is able to store constructs and their own genes for recovery later.<br /><br /> | ||
| - | Generating the primers for PCR extension takes only a few seconds each, and the information is returned to the user in the form of a PDF with a datasheet for each primer. In the near future it will also provide all the necessary protocols to complete your construct. As time goes by, more granular conrol over settings will become available. | + | Generating the primers for PCR extension takes only a few seconds each, and the information is returned to the user in the form of a PDF with a datasheet for each primer. In the near future it will also provide all the necessary protocols to complete your construct. As time goes by, more granular conrol over settings will become available.<br /><br /> |
| + | The algorithm for optimising primer length is still under development, so should not be entirely trusted until the software comes out of beta. When you find something that doesn't work, or could be improved, please <a href="mailto:info@gibthon.org?subject=Gison%20CD">email</a> the developer. | ||
<h4>How it works</h4> | <h4>How it works</h4> | ||
| - | The front is written in HTML and Javascript using the jQuery libraries. The backend is written in Python (with a tiny bit of php, which will hopefully disappear soon), connecting to a MySQL database. Melting temperature calculations and access to Entrez are done with the BioPython module, and Gibbs free energy/secondary structure calculations are done with UNAfold. | + | The front is written in HTML and Javascript using the jQuery libraries. The backend is written in Python (with a tiny bit of php, which will hopefully disappear soon), connecting to a MySQL database. Melting temperature calculations and access to Entrez are done with the BioPython module, and Gibbs free energy/secondary structure calculations are done with UNAfold. For more information on the tools used, please have a look <a href="http://www.gibthon.org/#about">here</a> |
</html> | </html> | ||
{{:Team:Cambridge/Templates/footerMinimal}} | {{:Team:Cambridge/Templates/footerMinimal}} | ||
Revision as of 15:40, 23 October 2010

Gibson Assembly

Gibthon Construct Designer
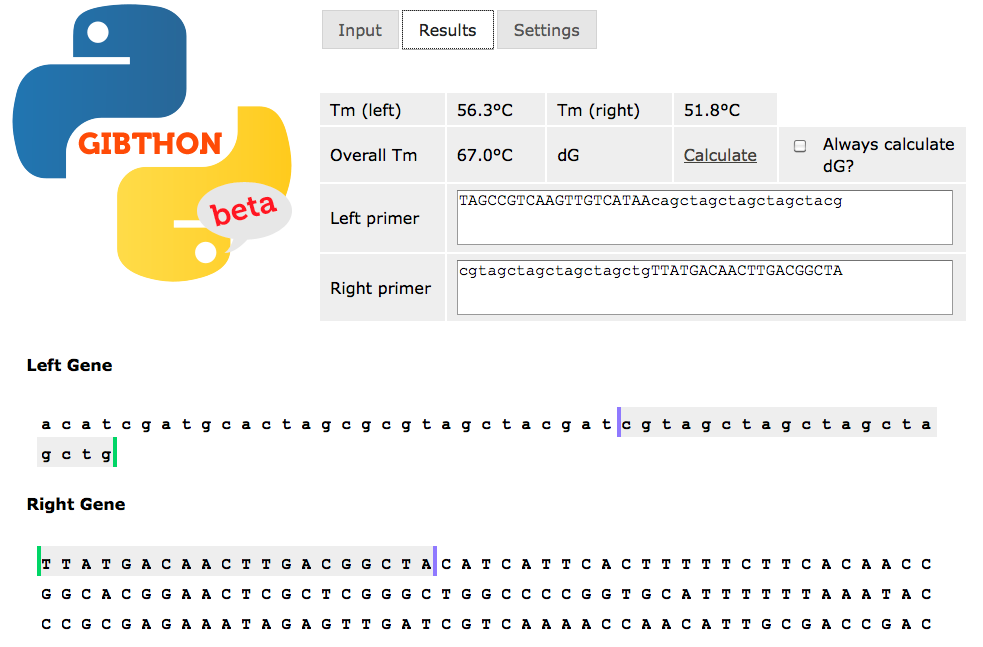
is a software tool under active development to aid with the design of primers for Gibson Assembly. It currently comes in two flavours - Gibthon Beta, and Gibthon Construct Designer Beta. The software is released under the BSD license, and the source code can be obtained for either by emailGibthon Beta - here
The more basic of the two tools, this will take as an input the sequences surrounding the join for which you wish to design primers. You can interactively adjust the length of the primers to tune the melting temperature of the DNA and check for secondary structures.The developer is currently focussing on GCD, but will still be supporting Gibthon - contact him here
Gibthon Construct Designer Beta - here
Gibthon's older brother, and much more powerful. With this, you can design your construct from its component genes and automatically generate the optimal primers. The tool has access to the Parts Registry Database, the NCBI's Nucleotide database, as well as accepting genbank formatted files from the user. The user is able to store constructs and their own genes for recovery later.Generating the primers for PCR extension takes only a few seconds each, and the information is returned to the user in the form of a PDF with a datasheet for each primer. In the near future it will also provide all the necessary protocols to complete your construct. As time goes by, more granular conrol over settings will become available.
The algorithm for optimising primer length is still under development, so should not be entirely trusted until the software comes out of beta. When you find something that doesn't work, or could be improved, please email the developer.
How it works
The front is written in HTML and Javascript using the jQuery libraries. The backend is written in Python (with a tiny bit of php, which will hopefully disappear soon), connecting to a MySQL database. Melting temperature calculations and access to Entrez are done with the BioPython module, and Gibbs free energy/secondary structure calculations are done with UNAfold. For more information on the tools used, please have a look here
 "
"