Team:Cambridge/Notebook/4
From 2010.igem.org
EmilyKnott (Talk | contribs) |
EmilyKnott (Talk | contribs) |
||
| (One intermediate revision not shown) | |||
| Line 23: | Line 23: | ||
Bill, Emily, Anja tried to add a prokaryotic RBS and promoter to the firefly luciferase already in the registry. They performed miniprep and restriction digest then ligation, but used incorrect restriction enzymes, such that we could not exclude religated plasmid backbones. | Bill, Emily, Anja tried to add a prokaryotic RBS and promoter to the firefly luciferase already in the registry. They performed miniprep and restriction digest then ligation, but used incorrect restriction enzymes, such that we could not exclude religated plasmid backbones. | ||
| + | |||
{{Team:Cambridge/Templates/Day|Day=Thursday}} | {{Team:Cambridge/Templates/Day|Day=Thursday}} | ||
{{:Team:Cambridge/Templates/rightpic|src=Cambridge-Glow.jpg}} | {{:Team:Cambridge/Templates/rightpic|src=Cambridge-Glow.jpg}} | ||
| Line 81: | Line 82: | ||
** The firely luciferase, and backbones, for miniprep if necessary | ** The firely luciferase, and backbones, for miniprep if necessary | ||
* + made up the 5mlsish of LB with pHK555/724 (Slock's strain) with 200ml LB | * + made up the 5mlsish of LB with pHK555/724 (Slock's strain) with 200ml LB | ||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 15:32, 27 October 2010
Contents |
Monday
We transformed some of the competent TOP10 cells we made the previous week with plasmid phK724, which was kindly sent to us by [http://departments.kings.edu/biology/lux/ Jim Slock]
We also placed E. coli containing phK555 in 5ml LB broth in a rotating 37°C incubator to grow up overnight.
In terms of dry work, we discussed what we wanted to have synthesised. We decided to dismiss PPDK (Pyruvate Orthophosphate Dikinase) and to include the Japanese firefly (Luciola cruciata) because of the ease with which we could change the colour of its emission.
Tuesday
We made cells from 4 different hns mutants: W3110 hns93-1, BW25113 Δhns::kan, W3110 hns-205::Tn10, GM230 hns-205::Tn10. This was prompted by a paper suggesting increased bioluminescence in strains with reduced hns. These cells were stored at -80°C for later use
We continued to discuss synthesis requirements
Wednesday
Hannah and Theo tested the previous day's preparations for competence. Unfortunately they plated out only 20ul, not enough to get sufficient colonies for an accurate perception of competence. Meanwhile, Peter and Paul transformed E.coli containing phK555 with phK724 and placed the cells in a 30C incubator. (V. fischeri lux proteins denature above this temperature)
Bill, Emily, Anja tried to add a prokaryotic RBS and promoter to the firefly luciferase already in the registry. They performed miniprep and restriction digest then ligation, but used incorrect restriction enzymes, such that we could not exclude religated plasmid backbones.
Thursday
Results
GM230 hns-205 ::Tn10 TetR (black) - somewhat competent
W3110 hns 93-1 (blue) - no transformants
BW25113 Δ hns ::kan (red) - somewhat
W3110 hns-205::Tn10 TetR - no tansformants
Theo's plan
- Miniprep plasmid DNA from luciferase and backbone transformed TOP10 following Qiagen protocol
- Digest luciferase Biobrick with: XbaI, PstI to produce two fragments:
- P end---old backbone---E--Xend
- Xend--luciferase---S--Pend (desired)
- Digest backbone Biobrick with SpeI, PstI to produce:
- Send--short fragment--Pend
- Pend---backbone--E--X--promoter--RBS--Send (desired)
- Run both digest products on a gel
- Extract suitable lengthed fragments from gel:
- Promoter + backbone should be 2153 bp
- Luciferase should be 1653 bp (its 3426 bp backbone should be discarded)
- Perform ligation
- S and X are compatible so we get
Pend---backbone--E--X--promoter--RBS--SXscar--luciferase---S--Pend - The two Pends will also ligate, producing a circular desired plasmid
- S and X are compatible so we get
- Transform competent TOP10 cells with ligation product
What happened
- In the morning we were a bit disconsolate because there was nothing visible on the plates containing phK724 and phK555, and our transformation tests for the variant hns strains were less successful than we had hoped.
- However our mood changed when Jim A told us we could not hope for many transformants with 20ul of strains that were not TOP10. We decided to retest the strains with 100 ul plated out.{{
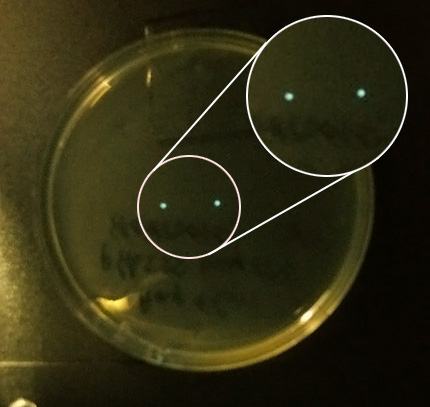
- AND we checked the 30 C incubator again and saw 2 colonies on one of the pHK555 plates, viewed under the camera they were clearly glowing so we gathered in the cold room and passed them around, as our eyes adapted we saw the colonies. We even saw a third glowing colony which we had not spotted on the plate.
Friday
Ligation appeared to have failed. Jim A suggests that we heat to 65 degrees to melt short DNAs after restriction digest, plus that we clean up the DNA with a spin column without gel.
PM
- Hannah and Theo transformed TOP10, red, and black containing phK555 with pHK724 using Jim Slock's EZ Competence buffer
- Anja, Emily drew out various things on petri dishes.
- Set off for trip to lake district
Saturday
We spent the day in the Lake District, a beautiful part of the UK 250 miles North of Cambridge.
Sunday
Went in to the lab on return: Found that:
- TOP10 containing 5/7 (Theo&Hannah, Friday) had been successfully transformed, glowing
- There were some colonies growing on ligation plate
- Logos were mostly dim, except at the edges (weekend incubation too long)
Theo and Peter:
- Redrew logos from *very* turbid liquid culture of pHK555/724
- Prepared overnight cultures of:
- Two hns strains (details?)
- The potential ligations
- The firely luciferase, and backbones, for miniprep if necessary
- + made up the 5mlsish of LB with pHK555/724 (Slock's strain) with 200ml LB
 "
"