Team:Cambridge/Gibson/Mechanism
From 2010.igem.org
(Difference between revisions)
(New page: {{:Team:Cambridge/Templates/headerMinimalprototype}} {{:Team:Cambridge/Templates/headerbar|colour=#96d446|title=Gibson Assembly: Introduction}} Gibson Assembly is a technique for assemblin...) |
|||
| Line 1: | Line 1: | ||
{{:Team:Cambridge/Templates/headerMinimalprototype}} | {{:Team:Cambridge/Templates/headerMinimalprototype}} | ||
| - | {{:Team:Cambridge/Templates/headerbar|colour=#96d446|title=Gibson Assembly: | + | {{:Team:Cambridge/Templates/headerbar|colour=#96d446|title=Gibson Assembly: Mechanism}} |
| - | + | ||
| + | Gibson Assembly is a means to join overlapping DNA sequences, technically it does not describe the way in which these sequences are created. However since this will be of importance to iGEM teams, we will briefly discuss this. | ||
| + | |||
| + | ===Creating overlapping DNA sequences=== | ||
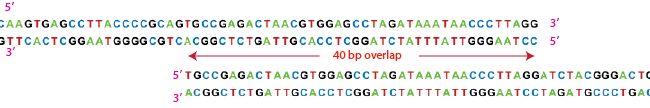
| + | Overlapping DNA sequences can be created by PCR. We can add twenty base-pairs to the end of a sequence by using a primer which runs as follows from 5' to 3'. | ||
| + | <pre> | ||
| + | 20 bp of sequence to add -> 20 bp of template to anneal to. | ||
| + | </pre> | ||
| + | By using two such primers we can add 20 bp of sequence A to sequence B and 20 bp of sequence B to sequence A. We are then ready to use Gibson Assembly. | ||
| + | |||
| + | [[Image:Cambridge-Gib1.png]] | ||
| + | |||
| + | == Gibson Assembly== | ||
| + | Gibson Assembly mix contains 3 enzymes: | ||
| + | * T5 exonuclease | ||
| + | * Phusion polymerase | ||
| + | * Taq ligase | ||
| + | [[Image:Cambridge-Gib2.png]] | ||
| + | [[Image:Cambridge-Gib3.png]] | ||
| + | [[Image:Cambridge-Gib4.png]] | ||
| + | [[Image:Cambridge-Gib5.png]] | ||
| + | [[Image:Cambridge-Gib6.png]] | ||
| + | [[Image:Cambridge-Gib7.png]] | ||
| + | [[Image:Cambridge-Gib8.png]] | ||
| + | [[Image:Cambridge-Gib9.png]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:Cambridge/Templates/footer}} | {{:Team:Cambridge/Templates/footer}} | ||
Revision as of 15:23, 18 September 2010

Gibson Assembly: Mechanism
Gibson Assembly is a means to join overlapping DNA sequences, technically it does not describe the way in which these sequences are created. However since this will be of importance to iGEM teams, we will briefly discuss this.
Creating overlapping DNA sequences
Overlapping DNA sequences can be created by PCR. We can add twenty base-pairs to the end of a sequence by using a primer which runs as follows from 5' to 3'.
20 bp of sequence to add -> 20 bp of template to anneal to.
By using two such primers we can add 20 bp of sequence A to sequence B and 20 bp of sequence B to sequence A. We are then ready to use Gibson Assembly.
Gibson Assembly
Gibson Assembly mix contains 3 enzymes:
- T5 exonuclease
- Phusion polymerase
- Taq ligase
 "
"