Team:Cambridge/Gibson/Introduction
From 2010.igem.org
(→Advantages) |
|||
| Line 2: | Line 2: | ||
{{:Team:Cambridge/Templates/headerbar|colour=#fb5c2b|title=Gibson Assembly: Introduction}} | {{:Team:Cambridge/Templates/headerbar|colour=#fb5c2b|title=Gibson Assembly: Introduction}} | ||
{{:Team:Cambridge/Templates/RightImage|image=GibsonAssembly.jpg|caption=Gibson Assembly joins any two lengths of DNA which have overlapping end regions}} | {{:Team:Cambridge/Templates/RightImage|image=GibsonAssembly.jpg|caption=Gibson Assembly joins any two lengths of DNA which have overlapping end regions}} | ||
| - | Gibson Assembly is a cutting-edge DNA ligation technique developed by Dan Gibson at JCVI in 2009 [http://www.nature.com/nmeth/journal/v6/n5/abs/nmeth.1318.html]. | + | '''Gibson Assembly''' is a cutting-edge DNA ligation technique developed by Dan Gibson at JCVI in 2009 [http://www.nature.com/nmeth/journal/v6/n5/abs/nmeth.1318.html]. |
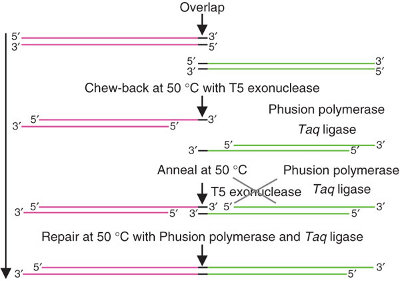
It uses three enzymes to ligate two or more sequences of DNA when they have overlapping end sequences at their joining point (~40bp). These overlapping regions can be easily added to the ends of any length of DNA by using PCR with primers which have added "flaps". Thus PCR followed by Gibson allows you to join any two blunt ended pieces of DNA. | It uses three enzymes to ligate two or more sequences of DNA when they have overlapping end sequences at their joining point (~40bp). These overlapping regions can be easily added to the ends of any length of DNA by using PCR with primers which have added "flaps". Thus PCR followed by Gibson allows you to join any two blunt ended pieces of DNA. | ||
Latest revision as of 20:47, 27 October 2010

Gibson Assembly is a cutting-edge DNA ligation technique developed by Dan Gibson at JCVI in 2009 [1].
It uses three enzymes to ligate two or more sequences of DNA when they have overlapping end sequences at their joining point (~40bp). These overlapping regions can be easily added to the ends of any length of DNA by using PCR with primers which have added "flaps". Thus PCR followed by Gibson allows you to join any two blunt ended pieces of DNA.
Advantages
1) No scar created - useful for fusion proteins and adding an RBS, where scars can be problematic.
2) Can re-ligate linear DNA into a circle - useful for site-directed mutagenesis
3) Any two blunt ended pieces of DNA can be joined, and DNA can be taken from any source which is accessible to PCR (for example, an organism's genome)
4) Although it is roughly the same speed as standard BioBrick assembly when ligating two fragments, Gibson is perfect for assembling multi part systems, since it takes the same amount of time to ligate n pieces of DNA together as it does for two pieces:
| Assembly Method | Number of steps to ligate N pieces of DNA |
| Standard Assembly | 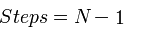
|
| Parallel Assembly | |
| Gibson Assembly |
Disadvantages
1) Greater degree of planning required as primers must be ordered in advance
2) More expensive than BioBrick assembly
3) PCR is tricky
Important:
The flexibility that this assembly method offers is a great thing. However, this does not mean it replaces the need for standardised prefixes and suffixes. The BioBrick prefix and suffix and the use of standard well-characterised vectors with standard primer sites are crucial for standardised iGEM parts. If you use Gibson Assembly it is important that you still add the prefix and suffix to your DNA; in fact sometimes Gibson Assembly is a useful way to do this (using as template an existing standard BioBrick).
 "
"