Team:Calgary/22 July 2010
From 2010.igem.org
Raidakhwaja (Talk | contribs) |
|||
| Line 1: | Line 1: | ||
{{CalgaryNotebookTemplate| | {{CalgaryNotebookTemplate| | ||
| - | |||
| - | |||
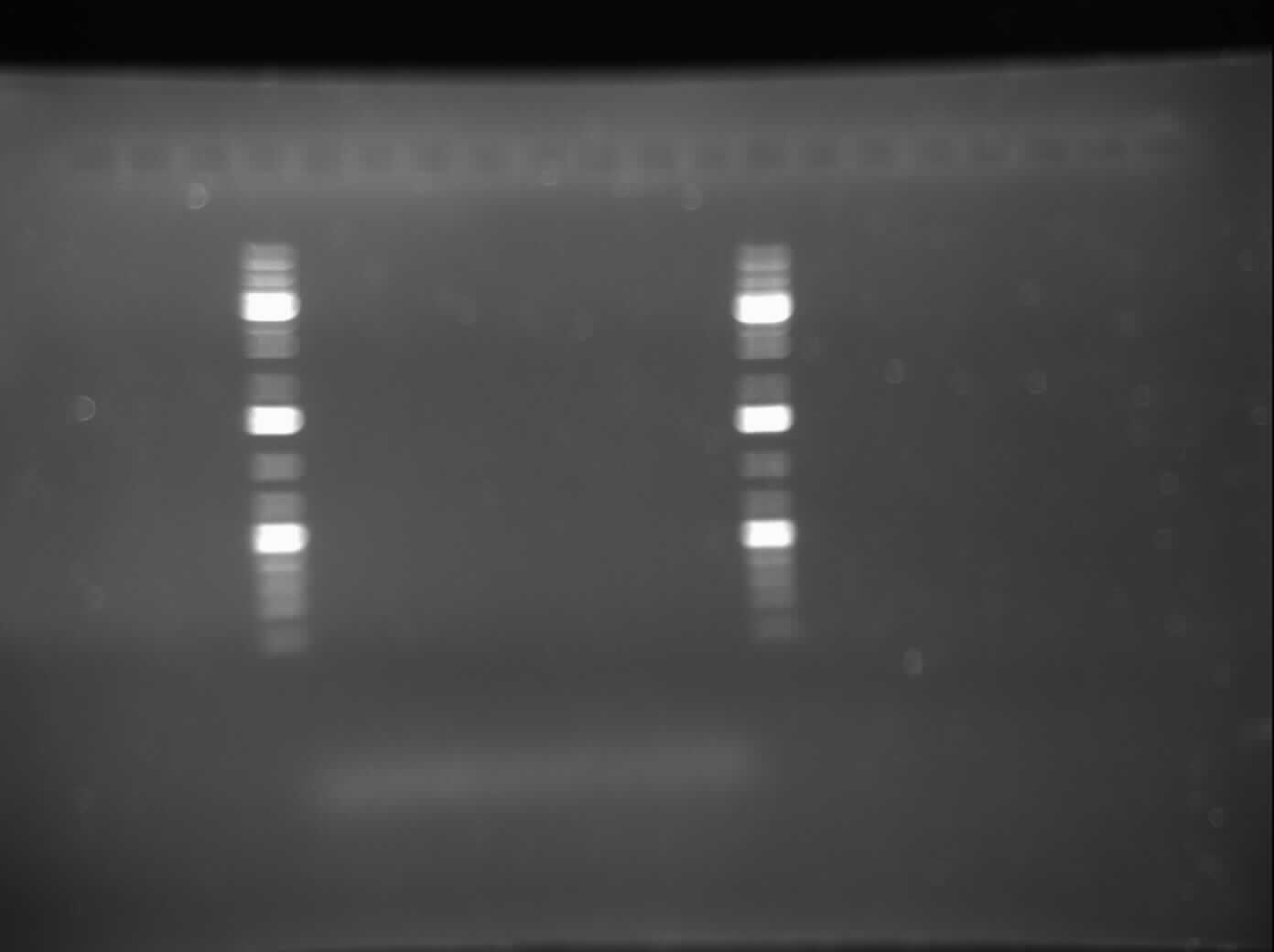
[[Image:07.21.2010 MalEPrimertesting.jpg|thumb|400px|Gel electrophoresis of Raida's PCR products: Mal-E Gene specific primers]] | [[Image:07.21.2010 MalEPrimertesting.jpg|thumb|400px|Gel electrophoresis of Raida's PCR products: Mal-E Gene specific primers]] | ||
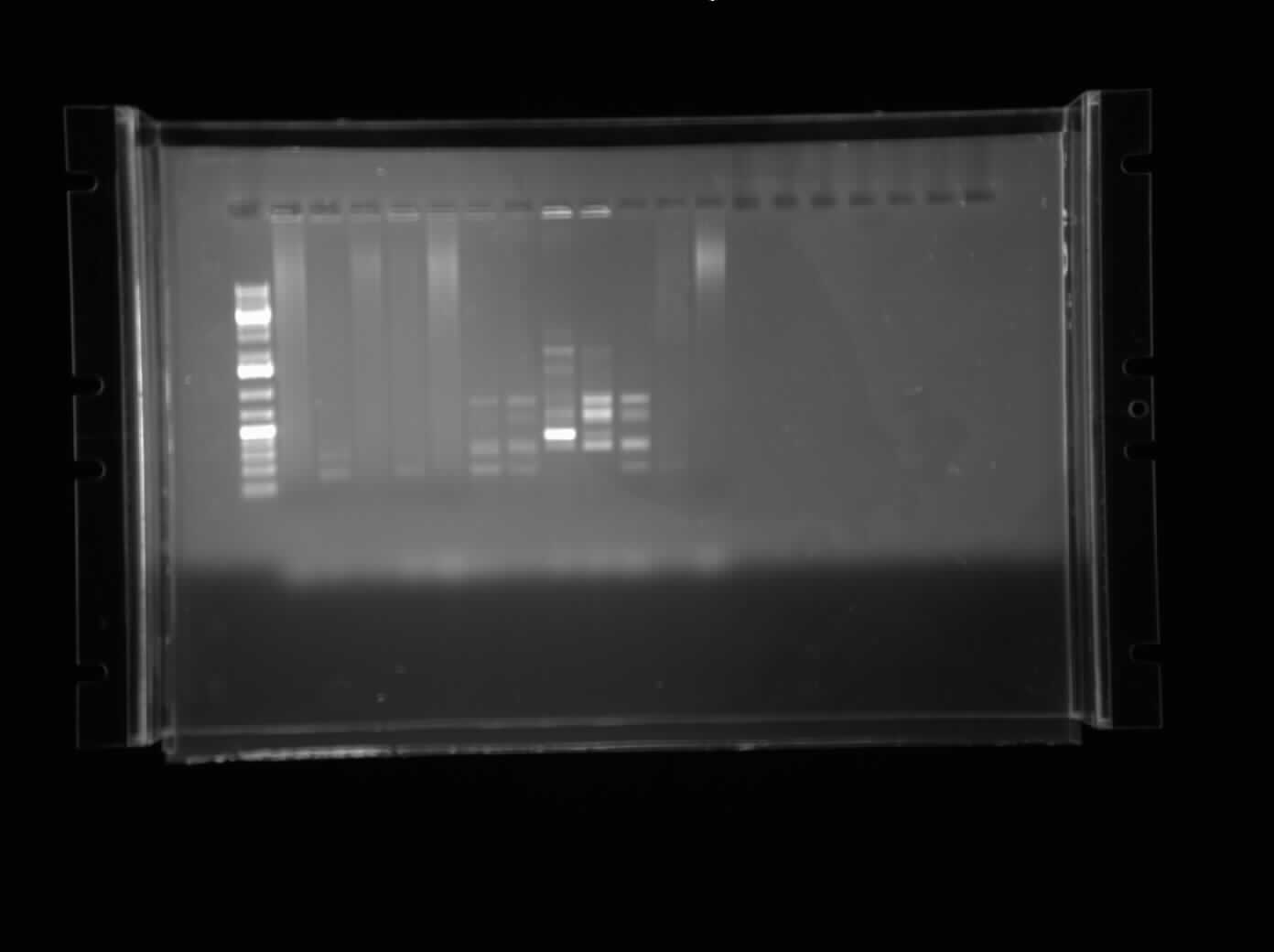
[[Image:07.22.2010 - CpxP Promoters Chris.jpg|thumb|400 px|Chris's Redone Gel Electrophoresis of the CpxP promoters in plasmid form. This time, the bands were very non-specific and a gradient PCR will be done tomorrow.]] | [[Image:07.22.2010 - CpxP Promoters Chris.jpg|thumb|400 px|Chris's Redone Gel Electrophoresis of the CpxP promoters in plasmid form. This time, the bands were very non-specific and a gradient PCR will be done tomorrow.]] | ||
| + | |||
| + | '''Thursday July 22, 2010''' | ||
| + | |||
<u>Himika</u> | <u>Himika</u> | ||
| Line 10: | Line 11: | ||
Today I miniprepped 10 colonies from the I0500-B0034 construction using Sigma-Aldrich kit. I also did a plasmid switch of I0500-B0034 from pSB1A3 to pSB1AC3. | Today I miniprepped 10 colonies from the I0500-B0034 construction using Sigma-Aldrich kit. I also did a plasmid switch of I0500-B0034 from pSB1A3 to pSB1AC3. | ||
| - | <u> Jeremy </u> | + | |
| + | <u>Jeremy</u> | ||
Today I did a colony PCR of K23900+I13507, K239000+I13504, K135000+I13507, K135000+I13504 and prepared an overnight of several colonies. I also re-ligated and transformed I0500+B0034 in pSB1AC3 plasmid. | Today I did a colony PCR of K23900+I13507, K239000+I13504, K135000+I13507, K135000+I13504 and prepared an overnight of several colonies. I also re-ligated and transformed I0500+B0034 in pSB1AC3 plasmid. | ||
| + | |||
<u>Chris</u> | <u>Chris</u> | ||
| Line 18: | Line 21: | ||
Today, I calculated the values of the enzymes we received from New England Biolabs. As well, I redid the gel of the colony PCR of the CpxP promoter with a 1.0% gel. This time, the DNA actually moved and the picture of the gel can be seen on the right. However, the bands were very non-specific. To optimize, I set up one gradient PCR with temperature of annealing step ranging from 53-58°C. | Today, I calculated the values of the enzymes we received from New England Biolabs. As well, I redid the gel of the colony PCR of the CpxP promoter with a 1.0% gel. This time, the DNA actually moved and the picture of the gel can be seen on the right. However, the bands were very non-specific. To optimize, I set up one gradient PCR with temperature of annealing step ranging from 53-58°C. | ||
| - | <u> Raida </u> | + | |
| + | <u>Raida</u> | ||
*Today, I did a gel electrophoresis of my PCR product from June 21, which was done to test the MalE primers. The results were negative as no bands were seen. This indicated that there was no amplication. I will do another PCR with the annealing temperature changed. I am also going to change the buffer because the picture came out blurry and I am predicting that it is because of the buffer. | *Today, I did a gel electrophoresis of my PCR product from June 21, which was done to test the MalE primers. The results were negative as no bands were seen. This indicated that there was no amplication. I will do another PCR with the annealing temperature changed. I am also going to change the buffer because the picture came out blurry and I am predicting that it is because of the buffer. | ||
| Line 25: | Line 29: | ||
* Near the end of the day, with the help of my supervisor, Henry, I set up and ran another PCR with the same DNA and the same primers. This time, the difference was that I used the Qiagen Master Mix. I was also suggested to change the annealing temperature to 52 degrees for 30 seconds instead of 55 degrees for 1 min as I did on June 21st. I will run a gel electrophoresis of my PCR product tomorrow to check whether the MalE gene specific primers annealed at the right location and amplified Mal-E gene. | * Near the end of the day, with the help of my supervisor, Henry, I set up and ran another PCR with the same DNA and the same primers. This time, the difference was that I used the Qiagen Master Mix. I was also suggested to change the annealing temperature to 52 degrees for 30 seconds instead of 55 degrees for 1 min as I did on June 21st. I will run a gel electrophoresis of my PCR product tomorrow to check whether the MalE gene specific primers annealed at the right location and amplified Mal-E gene. | ||
| + | |||
| + | <u>Patrick</u> | ||
| + | |||
| + | Paul checked up on the progress of our modelling project. The next task for us to determine what exactly happens to cells if they undergo excessive protein expression. A few questions to consider: what happens if the cell's overproduction of the stress response actually causes a stress to the cell itself? How do we go about correlating levels of fluorescence to expression levels? | ||
| + | |||
| + | Thus, I will proceed to look into some literature regarding any connection between optical density of Invitrogen TOP10 cells and its estimated cell concentration. | ||
}} | }} | ||
Revision as of 21:29, 23 July 2010

Thursday July 22, 2010
Himika
Today I miniprepped 10 colonies from the I0500-B0034 construction using Sigma-Aldrich kit. I also did a plasmid switch of I0500-B0034 from pSB1A3 to pSB1AC3.
Jeremy
Today I did a colony PCR of K23900+I13507, K239000+I13504, K135000+I13507, K135000+I13504 and prepared an overnight of several colonies. I also re-ligated and transformed I0500+B0034 in pSB1AC3 plasmid.
Chris
Today, I calculated the values of the enzymes we received from New England Biolabs. As well, I redid the gel of the colony PCR of the CpxP promoter with a 1.0% gel. This time, the DNA actually moved and the picture of the gel can be seen on the right. However, the bands were very non-specific. To optimize, I set up one gradient PCR with temperature of annealing step ranging from 53-58°C.
Raida
- Today, I did a gel electrophoresis of my PCR product from June 21, which was done to test the MalE primers. The results were negative as no bands were seen. This indicated that there was no amplication. I will do another PCR with the annealing temperature changed. I am also going to change the buffer because the picture came out blurry and I am predicting that it is because of the buffer.
Please refer to the image of the gel to the side.
- Near the end of the day, with the help of my supervisor, Henry, I set up and ran another PCR with the same DNA and the same primers. This time, the difference was that I used the Qiagen Master Mix. I was also suggested to change the annealing temperature to 52 degrees for 30 seconds instead of 55 degrees for 1 min as I did on June 21st. I will run a gel electrophoresis of my PCR product tomorrow to check whether the MalE gene specific primers annealed at the right location and amplified Mal-E gene.
Patrick
Paul checked up on the progress of our modelling project. The next task for us to determine what exactly happens to cells if they undergo excessive protein expression. A few questions to consider: what happens if the cell's overproduction of the stress response actually causes a stress to the cell itself? How do we go about correlating levels of fluorescence to expression levels?
Thus, I will proceed to look into some literature regarding any connection between optical density of Invitrogen TOP10 cells and its estimated cell concentration.
No notebook page exists for this date. Sorry! "
"