Team:Calgary/13 August 2010
From 2010.igem.org
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{CalgaryNotebookTemplate| | {{CalgaryNotebookTemplate| | ||
| - | + | Friday August 13, 2010| | |
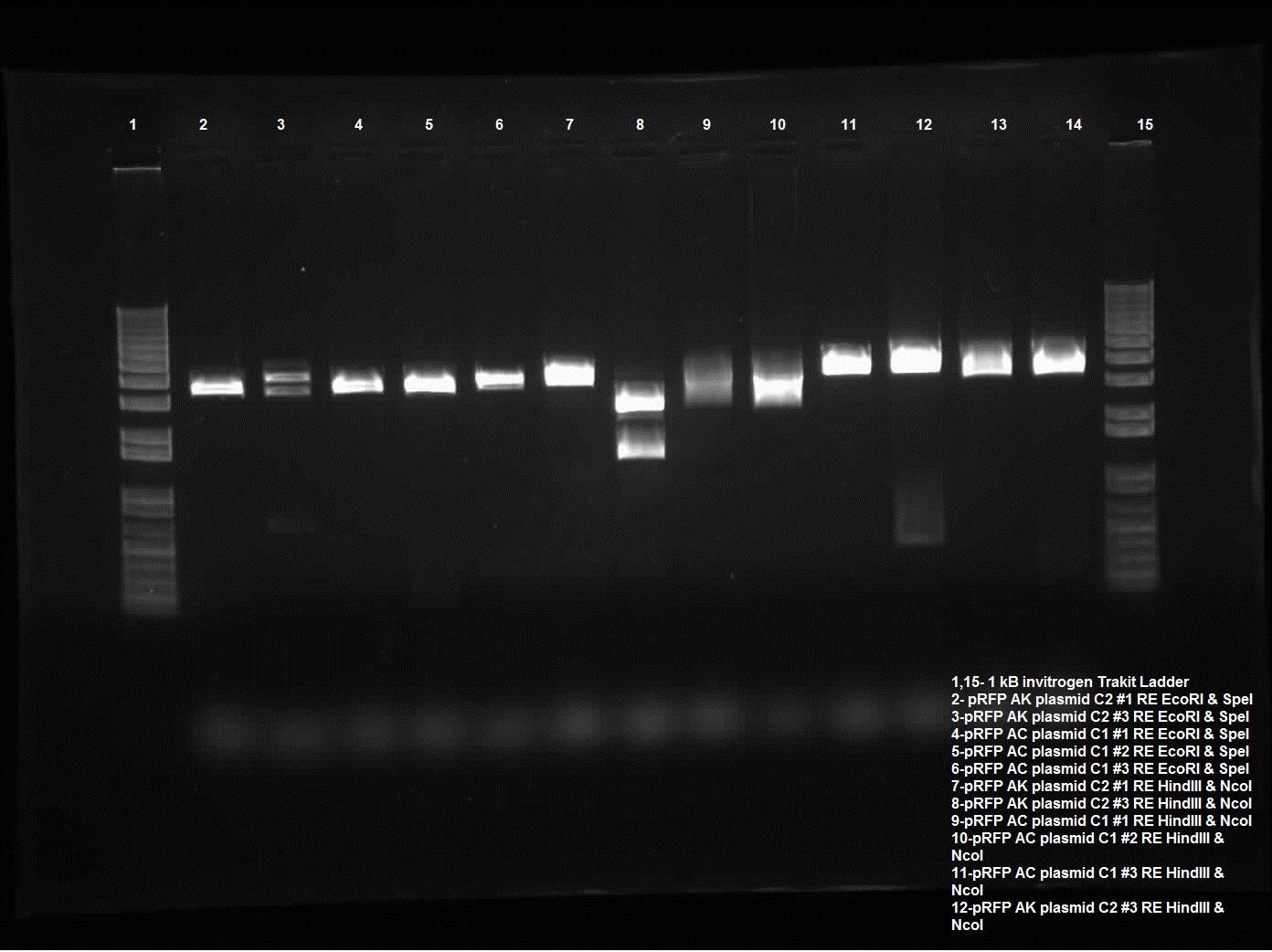
| + | [[Image:08.13.2010.Jeremy's pRFP RestrictionDigest.jpg|400px|thumb|Jeremy's Restriction Digest of pRFP in pSB1AK3 and pSB1AC 3 using EcoRI and SpeI, NcoI and HindIII, and PstI.]] | ||
| + | |||
| + | [[Image:IMG 0063.JPG|thumb|400px|Some of the laboratory materials given to us by VWR. Thanks to Ernesto Sanchez for these!]] | ||
''Friday the 13th...'' | ''Friday the 13th...'' | ||
<u>Himika</u> | <u>Himika</u> | ||
| - | |||
Today I started working on the presentation for aGEM which is on Sept 11 weekend. I layed out the presentation and added some templates. I thought about what I want it to look like. I have not put in any contents just yet. This will be done sometimes next week when everybody is present. I also got sequencing results back. It seems like the construction of I0500-B0034-MalE31 (C11) worked. This construct is currently in pSB1AC3. Although there is alignment, it looks like the tubes were mislabelled because the forward sequence lines up with the MalE31. MalE31 should have lined up with the reverse sequence. One of 2 things could have happened here: I chose the wrong enzymes while contructing or mislabelling. If I chose the wrong enzymes, the cells should not have grown in AC. They should have died.Therefore, it is likely that the tubes were labelled incorrectly. | Today I started working on the presentation for aGEM which is on Sept 11 weekend. I layed out the presentation and added some templates. I thought about what I want it to look like. I have not put in any contents just yet. This will be done sometimes next week when everybody is present. I also got sequencing results back. It seems like the construction of I0500-B0034-MalE31 (C11) worked. This construct is currently in pSB1AC3. Although there is alignment, it looks like the tubes were mislabelled because the forward sequence lines up with the MalE31. MalE31 should have lined up with the reverse sequence. One of 2 things could have happened here: I chose the wrong enzymes while contructing or mislabelling. If I chose the wrong enzymes, the cells should not have grown in AC. They should have died.Therefore, it is likely that the tubes were labelled incorrectly. | ||
| + | |||
<u>Jeremy</u> | <u>Jeremy</u> | ||
| - | Today I did a restriction enzyme of plasmid switch of pRFP into pSB1AK3 and pSB1AC3. The colonies that were chosen were AK C2 1 and 3 and AC C1 1 and 2 and AC C3 1. These were chosen because of the PstI sites cut yesterday, these ones looked the most promising. Today the restriction enzyme run for these included EcoRI and SpeI, HindIII and NcoI, and PstI. Only AK C2 3 behaved as I intended with two significant bands (the third being the uncut, this is most likely poor procedure with new NEB enzymes), two distinct lines one around ~1600-1700 kb and the other around 2500 kb. It was intended for an internal cut side so that a piece of pRFP was cut out however there was also a HindIII cut site in the plasmid. And lastly the PstI which cuts the internal cut site as well as the biobrick. The piece should be around 300 kb which is seen with pRFP AK C2 3. | + | Today I did a restriction enzyme of plasmid switch of pRFP into pSB1AK3 and pSB1AC3. The colonies that were chosen were AK C2 1 and 3 and AC C1 1 and 2 and AC C3 1. These were chosen because of the PstI sites cut yesterday, these ones looked the most promising. Today the restriction enzyme run for these included EcoRI and SpeI, HindIII and NcoI, and PstI. Only AK C2 3 behaved as I intended with two significant bands (the third being the uncut, this is most likely poor procedure with new NEB enzymes), two distinct lines one around ~1600-1700 kb and the other around 2500 kb. It was intended for an internal cut side so that a piece of pRFP was cut out however there was also a HindIII cut site in the plasmid. And lastly the PstI which cuts the internal cut site as well as the biobrick. The piece should be around 300 kb which is seen with pRFP AK C2 3. Gel was run on 0.8% using the Trackit ladder from Invitrogen. |
| + | |||
| + | |||
| + | <u>Chris</u> | ||
| + | Today, I started a restriction digest of different Registry parts pSB1AC3, pSB1AK3, I0500, I13507, CpxP promoter, and the ibpAB promoter. They were cut with either EcoRI and PstI or XbaI and PstI. I used the NEB EcoRI Buffer along with BSA as a 10x Buffer. As well, we received some laboratory materials from VWR for their sponsorship. I will do the ligations with the digests during the weekend. | ||
}} | }} | ||
Latest revision as of 05:01, 23 August 2010

Friday August 13, 2010
Friday the 13th...
Himika
Today I started working on the presentation for aGEM which is on Sept 11 weekend. I layed out the presentation and added some templates. I thought about what I want it to look like. I have not put in any contents just yet. This will be done sometimes next week when everybody is present. I also got sequencing results back. It seems like the construction of I0500-B0034-MalE31 (C11) worked. This construct is currently in pSB1AC3. Although there is alignment, it looks like the tubes were mislabelled because the forward sequence lines up with the MalE31. MalE31 should have lined up with the reverse sequence. One of 2 things could have happened here: I chose the wrong enzymes while contructing or mislabelling. If I chose the wrong enzymes, the cells should not have grown in AC. They should have died.Therefore, it is likely that the tubes were labelled incorrectly.
Jeremy
Today I did a restriction enzyme of plasmid switch of pRFP into pSB1AK3 and pSB1AC3. The colonies that were chosen were AK C2 1 and 3 and AC C1 1 and 2 and AC C3 1. These were chosen because of the PstI sites cut yesterday, these ones looked the most promising. Today the restriction enzyme run for these included EcoRI and SpeI, HindIII and NcoI, and PstI. Only AK C2 3 behaved as I intended with two significant bands (the third being the uncut, this is most likely poor procedure with new NEB enzymes), two distinct lines one around ~1600-1700 kb and the other around 2500 kb. It was intended for an internal cut side so that a piece of pRFP was cut out however there was also a HindIII cut site in the plasmid. And lastly the PstI which cuts the internal cut site as well as the biobrick. The piece should be around 300 kb which is seen with pRFP AK C2 3. Gel was run on 0.8% using the Trackit ladder from Invitrogen.
Chris
Today, I started a restriction digest of different Registry parts pSB1AC3, pSB1AK3, I0500, I13507, CpxP promoter, and the ibpAB promoter. They were cut with either EcoRI and PstI or XbaI and PstI. I used the NEB EcoRI Buffer along with BSA as a 10x Buffer. As well, we received some laboratory materials from VWR for their sponsorship. I will do the ligations with the digests during the weekend.
 "
"