|
Group Notebook for October 2010
Oct 26
@ Burkett Lab
Day before the freeze/deadline and we still cannot confirm the product's success or failure
through the colony PCR. We can thus not send out the product until after the due date, and thus
we will not be able to actually compete for a medal or anything. We do, however, still intend to
finish our part and submit it to the registry, as well as attending the Jamboree with our information
and present our work to anyone interested.
Oct 25
@ Burkett Lab
Colony PCR gave no results for the product, but the positive control gave no results as well.
We're too close to the wiki freeze deadline to be able to redo the colony PCR and still have
everything else done in preparation of getting our product sequenced in time for shipment.
The team decided to move forward and ship part
not sequenced and do the sequencing the week before the jamboree then update
the wiki after we return when it becomes unfrozen.
Oct 22
@ Burkett Lab
Patched colonies to LB & Amp, resuspended in 50 ul diH20 for colony pcr
Used 5 ul of pooled colonies & 5 ul of 10 to the -1 dilution in separate rxns.
2-5 ul Taq genomic DNA as a control
Oct 21
@ Burkett Lab
Digest of ligation rxns incubating for 2 hours @ 37 degrees C
Robert & Bernadette completed site directed mutation using the Stratagene protocol
3 rxns containing : 7.8 ul Reverse Mechanism, 7.4 ul Forward Mechanism
5 ul DNA, 5 ul Pfu turbo buffer, 1 ul dNTP, 1 ul Pfu turbo, 11.8 ul diH20
Added 1 ul DpNI to pcr rxns incubated at 37 degrees C for 1.5 hrs
Transform NEB-10 lamba cells with 5 ul of PCR/DpNI rxn. Select on Amp. 75 ug/ml
Incubated on ice, heat shock & plated
Got colonies from transformation


Oct 20
@ Burkett Lab
Bernadette weighed tubes and material to purify. 2 combined samples Plasmid psB1C3 weighed .4
Taq sample weight .16. Used Q1 Aquick Gel Extraction kit to purify.
Electrophoresis on purifications against 1 kb ladder.
Ligation reactions completed with a no Taq control and a no enzyme control
4 ul plasmid, 4 ul Taq, 1 ul buffer, 1 ul enzyme. No taq control also included 4 ul diH20
No enzyme control had 1 ul diH20. Incubated at 16 degrees C overnight
Oct 19
@ Burkett Lab
Bernadette completed digests. Promoter R0010 cut with EcoRI 1 ul and SpeI 1 ul
Buffer 4 5 ul, 10 ul DNA, 5 ul BSA (diluted 1:10) and 28 ul diH20. Also ran an uncut sample
with 30 ul diH20.
RBS B0034 cut with XbaI 1ul and PstI 1 ul also had uncut sample
Plasmid pSB1C3 and J04455 cut with EcoRI 1ul and PstI 1 ul also with uncut control
Ran gel and cut out plasmid and Taq to purify
Oct 18
@ Burkett Lab
Prepared plasmid mini prep B0015 tt, B0034 RBS, R0010 lacI promoter. Grew up cells
harvested parts. Used quick lysis kit.
Innoculated Taq into vector 5 ul LB media, 5 ul chlorophenocol resistant plasmid B0012
and B0011. 15 ul tetracycline, 25 ul kanamycin

Oct 14
@ Burkett Lab
Robert was testing our product from overlap against pcr conditions and possible errors.
Our last 4 gel electrophoresis of pcrs have raised some concerns about whether our new POLI
pstI site template is what it should be and if our primers are still good with it.
First thing we have done today "Blast" the primers against the original POLI template
to check for possible binding sites. There are a few around locations that could cause problems but
they are not closer than 100 bp to the single base mutation we made - thus shouldn't account
for the differences. Looking back at the recent pcrs, a temperature gradient hasn't been employed
against the new template yet, and we may need higher stringency in our new, bred template.
Robert set up pcr reaction for 5 reactions with varying annealing temperatures
to compare against the TAQ genomic POLI gene
Test 1 @ 58 degrees C, Test 2 60 degrees, Test 3 62 degrees, Test 4 64 degrees, Test 5 66 degrees
Results of our overlap had given us something of the right size, but not our product or the POLI
gene anymore. As the Overlap PCR has given no good results after seven attempts, and the seventh was
a counterfeit of the right size just to make it difficult to tell if the Overlap was successful, we
will be attempting a different approach. In order to make sure that we have something to submit, even
if not our intended product, we will be working on placing a POLI gene with the BB-prefix and suffix
in the required plasmid as well as trying a new method for the single base mutation. The new method
mimics a kit (Stratagene) designed to be used while a gene is in a plamid already. This procedure first
copies the whole plasmid on the inside and outside (with the mutation from the primer) and then uses dpnI
to remove the methylated original plamid, leaving only the intended product already in the plasmid.
Modified template vs. Genomic in same reaction:

Oct 13
@ Burkett Lab
results for POLI - PSTI:
 odd...
odd...
Oct 12
@ Burkett Lab
Robert ran another pcr with 2 tests: diH20 25.5 ul, buffer 10 ul, dNTP 1 ul, DMSO 2.5, Fwd primer 2.5
Rv primer 2.5 ul, Pol I 5 ul, PFU 1 ul. The second test was the same except 10ul POLI and 20.5 ul diH20
pcr ran with an annealing temp of 58.5 degrees C.
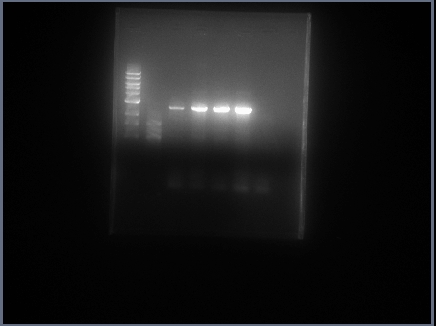
got good results, ran PCR with POLI - PSTI.
Bernadette ran a gel on Promoter, Double Terminator and RBS digests cut and uncut.
RBS and Double Terminator looked fine. Bernadette cut out from gel - will redo Promoter.

Oct 11
@ Burkett Lab
Robert's pcr gel was blank except for the control lane - He'll rerun tomorrow
Bernadette did RBS digest w/ 15 ul DNA, XbaI 1 ul, PstI 1 ul, BSA .5, Buffer 4 @ 5 ul, diH20 27.5. Also
an uncut control with 29.5 ul diH20. Promoter digest was identical except used SpeI 1 ul and Pst I 1 ul -
Also an uncut control. Double terminator Digest the same except EcoRI 1 ul and XbaI 1 ul w/uncut control.
Oct 7
@ Burkett Lab
Bernadette & Robert
Gel results were not what was expected for amplification, but the product is good after gel purification
Robert tested PCR w/ original TAQ to get optimal conditions. Tested different gradients of DMSO/temps.
Bernadette ran gel on double term, RBS & Promoter

Oct 6
@ Burkett Lab
Robert's gel results from pcr (58 degrees C, 2.5% DMSO & 61 degrees, 2.5% DMSO) yield results that match
TAQ poli in size. Part extracted & purified. Next..Amplification through cloning into a cell and PCR

|  "
"





 odd...
odd... 


