Team:BCCS-Bristol/Modelling/BSIM/Results
From 2010.igem.org
| Line 7: | Line 7: | ||
==Motility in a Gel Matrix== | ==Motility in a Gel Matrix== | ||
| + | |||
| + | A question that we wanted to analyze was how the 'run-and-tumble' motion of ''E. coli'' is affected by them being embedded in a gel. The gel is not completely solid, rather it is composed of fluid filled voids and strands of sugary gellan. We devised a simulation to track the movement of ''E. coli'' in a segment of a gellan bead. | ||
| + | |||
| + | ===Analysing the Gel Structure=== | ||
| + | |||
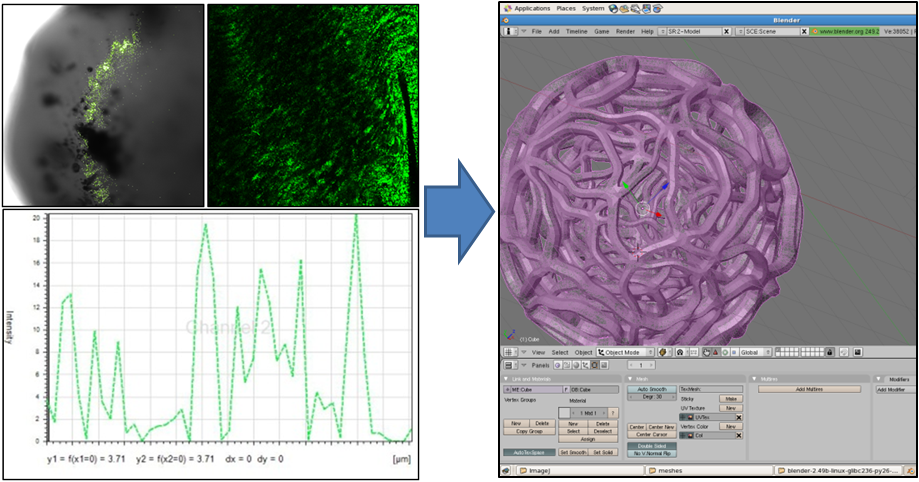
| + | Our first step was to find out what the inside of the gel looked like on a microscale. Using a high-magnification confocal microscope we mapped the distribution of E. coli in the gel matrix to give us a sense of the density of the gel. By looking at the fluorescence of | ||
| + | |||
| + | [[Image:Mesh_composite.png|center|450px|Confocal data]] | ||
Revision as of 10:01, 27 October 2010
iGEM 2010
Contents |
Results: Overview
On this page we have presented a few examples of what can be done with the environmental modelling package in BSim 2010. These examples illustrate the three core featuers of the package, namely importing 3D meshes, generating and processing arbitrarily shaped chemical fields and collisions.
Motility in a Gel Matrix
A question that we wanted to analyze was how the 'run-and-tumble' motion of E. coli is affected by them being embedded in a gel. The gel is not completely solid, rather it is composed of fluid filled voids and strands of sugary gellan. We devised a simulation to track the movement of E. coli in a segment of a gellan bead.
Analysing the Gel Structure
Our first step was to find out what the inside of the gel looked like on a microscale. Using a high-magnification confocal microscope we mapped the distribution of E. coli in the gel matrix to give us a sense of the density of the gel. By looking at the fluorescence of
Two simulations look at how motile agrEcoli move through a volume of the bead. The average pore diameter is 15um. In the first simulation the gel is hole, in the second the bonds between sugars have broken down creating a less coherent gel. Using BSim we tracked the movement of the agrEcoli to see how this affected their movement.
Tracking Movement
As an illustration of the fact that one can take quantitative data out of BSim rather than just visualizations we've calculated the average distance moved by a sample of agrEcoli in the two gels. It is greater in the intact gel matrix, one might speculate that this is because the continuous voids in this gel offer less opportunity for the bacteria to become tangled in the gel.
Whole Gel Matrix

Degraded Gel Matrix

Simple Diffusion
Simple illustration of chemical field diffusing through two different media, the aqueous environment surrounding the gel strand and the gel itself. The source of the chemical is a contact at the bottom of the simulation volume. The agrEcoli bacteria are shown changing color as concentration changes to represent expression of rfp/gfp ratio.
 "
"