Project/Lac1/AraC Promoter/
From 2010.igem.org
| Line 9: | Line 9: | ||
=== Sequence Developement and Annotation === | === Sequence Developement and Annotation === | ||
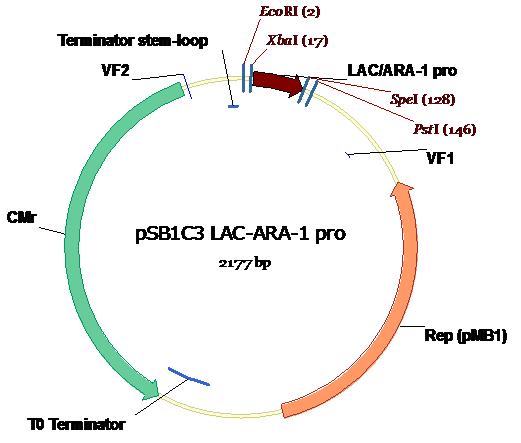
| + | The Lac1/AraC promoter (below) as submitted to The Registry in the Standard Assembly PSB1C3 plasmid backbone (exhibiting chloramphenicol resistance) with appropriate annotations of Standard Assembly restriction sites as well as key plasmid features. Illustration courtesy Dr Marco Weinberg [2]. | ||
| + | [[Image:LacAraC_in_PSB1C3.JPG]] | ||
| + | |||
| + | |||
| + | |||
<br /> | <br /> | ||
=== Testing and Validation === | === Testing and Validation === | ||
| Line 15: | Line 20: | ||
[1] Lutz R. & Bujard H. (1997), Independent and tight regulation of transcriptional units in Escherchia coli via the LacR/O, the TetR/O and AraC/I_1- I_2 regulatory elements. Nucleic Acids Research 25: 1203-1210. | [1] Lutz R. & Bujard H. (1997), Independent and tight regulation of transcriptional units in Escherchia coli via the LacR/O, the TetR/O and AraC/I_1- I_2 regulatory elements. Nucleic Acids Research 25: 1203-1210. | ||
<br /> | <br /> | ||
| + | [2] Marco Weinberg, PhD. Antiviral Gene Therapy Research Unit, Department of Molecular Medicine and Haematology, University of the Witwatersrand Medical School, 7 York Rd. Parktown 2193 SOUTH AFRICA, marc.weinberg@wits.ac.za. | ||
Revision as of 20:32, 14 October 2010
Contents |
Lac1/AraC IPTG-Inducible Promoter
Background
While the string of open reading frames lie mute within Lacto-sense, the trigger that errs it into motion upon sighting IPTG, the chosen proxy for HPV presence, is the Lac1/AraC promoter. This fusion promoter recruits elements from promoters belonging to both the Lactose and Arabinose operons to create a synthetic IPTG (or appropriate Arabinose isomer) -inducible promoter. The promoter was synthesized as described by Lutz and Bujard [1] via annealed primers (that share complementary sequences of overlap) and their subsequent elongation via PCR growth.
Polymerase binding is inhibited by the presence of the constitutively-produced AraC protein which binds to a specific sequence upstream of the -33 hexamer. DNA-AraC decoupling is brought about due to the presence of the Arabinose sugar, or a suitable isomer such as IPTG, which creates a conformational shift in the protein thus decreasing its DNA binding affinity. Thus, addition of exogenous Arabinose or IPTG will induce polymerase engagement with the promoter and subsequent transcription of the downstream open reading frames. Due to the duality in activation of both Arabinose and IPTG, promoter activity can be activated to differing degrees by the selective addition of either or both inducers and hence activity can be finely regulated via appropriate dose concentration administration.
Sequence Developement and Annotation
The Lac1/AraC promoter (below) as submitted to The Registry in the Standard Assembly PSB1C3 plasmid backbone (exhibiting chloramphenicol resistance) with appropriate annotations of Standard Assembly restriction sites as well as key plasmid features. Illustration courtesy Dr Marco Weinberg [2].
Testing and Validation
References
[1] Lutz R. & Bujard H. (1997), Independent and tight regulation of transcriptional units in Escherchia coli via the LacR/O, the TetR/O and AraC/I_1- I_2 regulatory elements. Nucleic Acids Research 25: 1203-1210.
[2] Marco Weinberg, PhD. Antiviral Gene Therapy Research Unit, Department of Molecular Medicine and Haematology, University of the Witwatersrand Medical School, 7 York Rd. Parktown 2193 SOUTH AFRICA, marc.weinberg@wits.ac.za.
 "
"