Filamentous Cells
From 2010.igem.org
(Difference between revisions)
RachelBoyd (Talk | contribs) |
RachelBoyd (Talk | contribs) |
||
| Line 1: | Line 1: | ||
| + | {{Team:Newcastle/mainbanner}} | ||
'''Filamentous Cells''' | '''Filamentous Cells''' | ||
{| | {| | ||
Revision as of 14:18, 30 May 2010

| |||||||||||||
| |||||||||||||
Filamentous Cells
| SOS response is believe to be a universal bacteria phenomenon first studied in E.coli -LexA, recA |
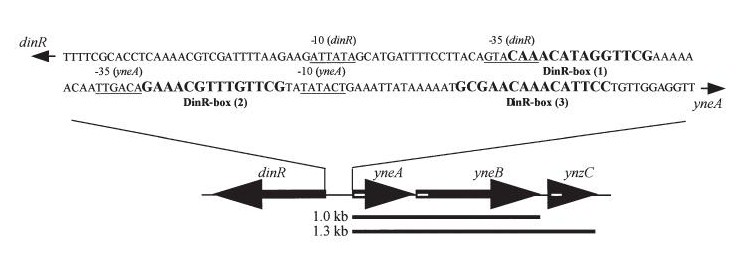
| In Bacillus subtillis (gram positive) dinR protein is homologous to lexA (Repressor of din-damage inducible genes).din genes include uvrA, uvrB, dinB, dinC dinR and recA. DNA damage inhibits cell division. |
| Wild type Bacillus subtillis |
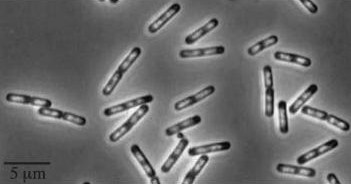
|
| dinR KO |
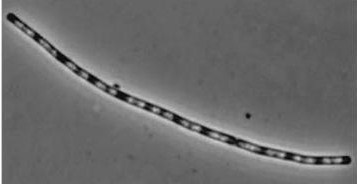
|
| dinR KO mutant over expressed the divergent (opposite direction) transcript for YneA, YneB and YnzC. These genes form the SOS regulon (recA independent SOS response) |
|
| YneA suppressed in wt without SOS induction |
| Expression of YneA from ITPG controlled promoter in wt leads to elongation. |
| Disruption of YneA in SOS response leads to reduced elongation. Altering YneB and YnzC expression does not affect cell morphology. |
| Double mutant (dinR/YneA) |
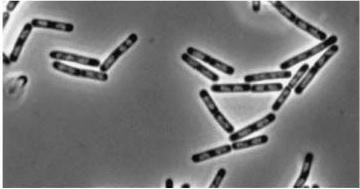
|
| YneA protein required to suppress cell division. Not chromosome replication or segregation. |
| FtsZ is important for bacterial cell division forming a ring structure at the division site by polymerising
assembling other proteins necessary for division at the site. |
| FtsZ localises to the cell division cycle unless dinR is disrupted or YneA is being induced. |
| YneA suppresses FtsZ ring formation- no proven direct interaction by two-hybrid. |
| Filamentous cells less colony formation. |
| YneA expression via the inactivation of dinR by Rec A is important. |
| Kawai, Y., Moriya, S., & Ogasawara, N. (2003). Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Molecular microbiology, 47(4), 1113-22. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12581363. |
 "
"