BBFRFC66
From 2010.igem.org
BBF RFC 66: A RESTful API for Supporting Automated BioBrick Model Assembly
Steyn, J. S.; Boyd, R. M; Essa, Y.; Hall, P.; Koh, A.; Sheth, H.; Tsu, D; Woodhouse, S; Hallinan, J.; Wipat, A.; Pocock, M. R.
23 September 2010
Contents |
Purpose
Constructing simulatable models for BioBricks by hand is a complex and time-consuming task. The time taken could be reduced by using Computer Aided Design (CAD) tools to aid in designing models, but these tools need to be augmented with domain-specific knowledge. Here we propose a standard for a RESTful (Richardson, 2007) API which facilitates the discovery and publication of models of functional biological units. This API is designed to produce parts models which can be automatically combined into complete, simulatable models of entire systems.
Relation to other BBF RFCs
BBF RFC 66 does not update or replace any earlier BBF RFC.
Copyright Notice
Copyright (C) The BioBricks Foundation (2010). All Rights Reserved.
Motivation
Computational simulation is an integral part of the BioBrick design process. However, given that realistic systems often involve multiple BioBricks, and that each BioBrick may contain multiple functional components (Knight, 2003), constructing simulatable models of biological systems can be a daunting task. Genetic circuits can be designed either manually, using CAD tools (e.g. Chandran, Bergmann & Sauro, 2009), or automatically (Rodrigo, Carrera & Jaramillo, 2007). To support these approaches several researchers have proposed methods for the bottom-up assembly of BioBricks (Marchisio & Stelling, 2008, Cooling et al., 2010). To achieve this task these tools require access to a library of models for individual parts, and enough additional information about these parts to allow the user to quickly identify valid combinations which allow the software to combine model fragments into larger models. Here we describe a RESTful service API for querying and fetching parts, model fragments for these parts, and combinatorial information about these parts, to support both automated and human-directed construction of complex models.
Terminology and Typography
Text appearing in fixed-width font denote parts of the RESTful API, such as data-types and operations. Text appearing in italic font indicate names and identifiers. The words ‘part’, ‘device’ and ‘system’ in standard font have the usual BioBrick meaning.
Data Model
The data model is hierarchical (Fig. 1). At the top level, an assembly bundles parts, connections and other assemblies together, producing re-usable, simulatable systems. Parts represent biological primitives and their associated models, including any publicly visible variables. Connections join parts together via their variables.
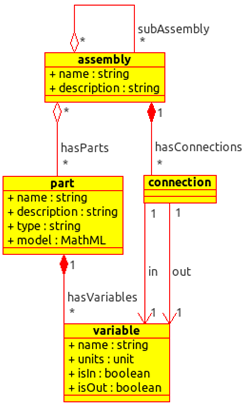 Figure 1. Data model class diagram
Data model types
There are four data types in the data model. Their structure is described in this section.
• Assembly: A device or system that can be independently simulated
An Assembly has the following attributes:
• name: The assembly name.
• description: An optional human-readable description of the assembly.
• subAssembly: Zero or more assemblies of which this assembly is composed.
• hasParts: The parts assembled by this assembly. Individual Parts may be assembled into any number of assemblies.
• hasConnections: The connections that wire parts together. For example, the POPs output of a promoter may be wired to the POPs input of a coding sequence.
• Part: An encapsulation of a quantified entity
A Part consists of:
• name: The part name.
• description: An optional human-readable description of the part.
• type: The biological type of the part, such as Promoter, CDS, RBS. This SHOULD come from an appropriate controlled vocabulary.
• model: A MathML block describing the relationships between the variables of this part.
• hasVariables: The variables exposed.
• Variable: An exposed quantity
A Variable has:
• name: No two variables within the same part may share the same name. However, variables from different parts may have the same name.
• units: The physical units of the variable. Defaults to dimensionless.
• isIn: A flag to indicate if this is an input variable for the part. Defaults to false
• isOut: A flag to indicate if this is an output variable for the part. Defaults to false
• Connection: A binding of an out-variable from one component to an in-variable of another component
• in: the in-variable to connect
• out: the out-variable to connect
Data model well-formedness constraints
For data to be well-formed, it must both conform to the data model described above, and additionally adhere to the following constraints:
• assembly.name (scope): each name uniquely identifies a single assembly. Assemblies MUST NOT share names. These MAY be re-used between entities of different types, for example between an assembly.name and a part.name.
• part.name (scope): each name uniquely identifies a single part. Parts MUST NOT share names. These MAY be re-used between entities of different types, for example between a part.name and a variable.name.
• part.hasVariables and part.model (scope): Every name of every variable of a part MUST appear in the model of that part.
• Variable.name (scope): Every name of every variable of a part MUST be unique within that part. Variables of different parts MAY (and in some circumstances MUST) use the same name.
• connection.in and connection.out (flags): These MUST refer to variables with the isIn and isOut flag set to true, respectively.
• connection.in and connection.out (scope): These MUST refer to variables that are within parts that are within the assembly holding the connection.
Data Rendering
Individual implementers are free to chose from one of several renderings of the data model. These include XML and JavaScript Object Notation (JSON).
To keep per-fetch costs low, this specification states what data MUST be present in a particular rendering. In particular, it states what MUST be directly embedded in-line as a value, and what MUST be included by reference to a name.
Field Type Encoding
Assembly
name string value
description string value
subAssembly List of assembly reference ${subAssembly.name}
hasPart List of part reference ${hasParts.name}
hasConnections List of connection value
Part
name string value
description string value
type string(cv) value
model MathML value
hasVariables List of variable value
Connection
in variable reference ${in.name}
out variable reference ${out.name}
Variable
name string value
units string(cv) value
isIn boolean value
isOut boolean value
Component Assembly
A CAD client is responsible for allowing the user to combine parts and assemblies to build a model of the system of interest. This is achieved by binding all unbound variables. A variable is unbound if it is not referred to by any connection. To ensure that the model can be simulated, the CAD program will need to bind each unbound in-variable to either an unbound out-variable, or to a constant value. For example, if an assembly modeling the Lac operon exposes in-variables named lac_repressor and inactivated_lac_repressor, these variables must be bound either to out-variables giving the levels of these molecules, or to suitable constant values.
Resources
REST is based around resources, identified by URLs. Given, for example, a base URL called BASE, the following resources MUST be provided by any implementation of this API: BASE/parts/ representing all parts; BASE/parts/${name} for each part; BASE/assemblies/ representing all assemblies; and BASE/assemblies/${name} for each assembly. For example, a server located at 'http://myuni.ac.uk/modelrepository' which published an assembly BA_K302012, would be exposed as: 'http://myuni.ac.uk/modelrepository/assemblies/BA_K302012'.
URL Description
parts/ All parts
Operation Result Example
GET The names of all parts Response: ['generic_promoter', 'Pspac', 'Pveg']
POST Create a new part Query: {
Figure 1. Data model class diagram
Data model types
There are four data types in the data model. Their structure is described in this section.
• Assembly: A device or system that can be independently simulated
An Assembly has the following attributes:
• name: The assembly name.
• description: An optional human-readable description of the assembly.
• subAssembly: Zero or more assemblies of which this assembly is composed.
• hasParts: The parts assembled by this assembly. Individual Parts may be assembled into any number of assemblies.
• hasConnections: The connections that wire parts together. For example, the POPs output of a promoter may be wired to the POPs input of a coding sequence.
• Part: An encapsulation of a quantified entity
A Part consists of:
• name: The part name.
• description: An optional human-readable description of the part.
• type: The biological type of the part, such as Promoter, CDS, RBS. This SHOULD come from an appropriate controlled vocabulary.
• model: A MathML block describing the relationships between the variables of this part.
• hasVariables: The variables exposed.
• Variable: An exposed quantity
A Variable has:
• name: No two variables within the same part may share the same name. However, variables from different parts may have the same name.
• units: The physical units of the variable. Defaults to dimensionless.
• isIn: A flag to indicate if this is an input variable for the part. Defaults to false
• isOut: A flag to indicate if this is an output variable for the part. Defaults to false
• Connection: A binding of an out-variable from one component to an in-variable of another component
• in: the in-variable to connect
• out: the out-variable to connect
Data model well-formedness constraints
For data to be well-formed, it must both conform to the data model described above, and additionally adhere to the following constraints:
• assembly.name (scope): each name uniquely identifies a single assembly. Assemblies MUST NOT share names. These MAY be re-used between entities of different types, for example between an assembly.name and a part.name.
• part.name (scope): each name uniquely identifies a single part. Parts MUST NOT share names. These MAY be re-used between entities of different types, for example between a part.name and a variable.name.
• part.hasVariables and part.model (scope): Every name of every variable of a part MUST appear in the model of that part.
• Variable.name (scope): Every name of every variable of a part MUST be unique within that part. Variables of different parts MAY (and in some circumstances MUST) use the same name.
• connection.in and connection.out (flags): These MUST refer to variables with the isIn and isOut flag set to true, respectively.
• connection.in and connection.out (scope): These MUST refer to variables that are within parts that are within the assembly holding the connection.
Data Rendering
Individual implementers are free to chose from one of several renderings of the data model. These include XML and JavaScript Object Notation (JSON).
To keep per-fetch costs low, this specification states what data MUST be present in a particular rendering. In particular, it states what MUST be directly embedded in-line as a value, and what MUST be included by reference to a name.
Field Type Encoding
Assembly
name string value
description string value
subAssembly List of assembly reference ${subAssembly.name}
hasPart List of part reference ${hasParts.name}
hasConnections List of connection value
Part
name string value
description string value
type string(cv) value
model MathML value
hasVariables List of variable value
Connection
in variable reference ${in.name}
out variable reference ${out.name}
Variable
name string value
units string(cv) value
isIn boolean value
isOut boolean value
Component Assembly
A CAD client is responsible for allowing the user to combine parts and assemblies to build a model of the system of interest. This is achieved by binding all unbound variables. A variable is unbound if it is not referred to by any connection. To ensure that the model can be simulated, the CAD program will need to bind each unbound in-variable to either an unbound out-variable, or to a constant value. For example, if an assembly modeling the Lac operon exposes in-variables named lac_repressor and inactivated_lac_repressor, these variables must be bound either to out-variables giving the levels of these molecules, or to suitable constant values.
Resources
REST is based around resources, identified by URLs. Given, for example, a base URL called BASE, the following resources MUST be provided by any implementation of this API: BASE/parts/ representing all parts; BASE/parts/${name} for each part; BASE/assemblies/ representing all assemblies; and BASE/assemblies/${name} for each assembly. For example, a server located at 'http://myuni.ac.uk/modelrepository' which published an assembly BA_K302012, would be exposed as: 'http://myuni.ac.uk/modelrepository/assemblies/BA_K302012'.
URL Description
parts/ All parts
Operation Result Example
GET The names of all parts Response: ['generic_promoter', 'Pspac', 'Pveg']
POST Create a new part Query: {
name: 'Plac',
description: 'Lac Operon promoter',
type: 'promoter',
model: '…', hasVariables: [{
name: 'RNA_polymerase',
isIn: true }, {
name: 'inact_lac_repressor',
isIn: true }, {
name: 'POPs',
isOut: true }]}
Result: parts/Placcreated
parts/ ${name} A part Operation Result Example GET Fetch the part Response: { name: '${name}',
… }
assemblies/ All assemblies Operation Result Example GET The names of all assemblies Response: ['Lac Operon', 'Bba_K302012'] POST Create a newassembly Query: {
name: 'BBA_K302012',
description: 'Filamentous cell phenotype',
subAssembly: [ 'BBA_K302003', 'BBA_K302005'],
hasConnections = [{
in: 'BBA_K302003.POPs',
out: 'BBA_K302005.POPs'} ]
Result: assemblies/BBA_K302012created
assemblies/ ${name} An assembly Operation Result Example GET Fetch an assembly Response: { name: '${name}', … }
Authors' Contact Information Jannetta S. Steyn: jannetta.steyn@ncl.ac.uk Rachel May Boyd: rachel.boyd@ncl.ac.uk Younus Essa: younus.essa@ncl.ac.uk Phil Hall: phillip.hall@ncl.ac.uk Alan Koh: s.c.a.koh@ncl.ac.uk Harsh Sheth: harsh.sheth@ncl.ac.uk Deena Tsu: deena.tsu@ncl.ac.uk Steven Woodhouse: steven.woodhouse@ncl.ac.uk Anil Wipat: anil.wipat@ncl.ac.uk Jennifer Hallinan: j.s.hallinan@ncl.ac.uk Matthew Pocock: matthew.pocock@ncl.ac.uk
References Chandran, D., Bergmann, F. T. & Sauro, H. M. (2009). TinkerCell: modular CAD tool for synthetic biology. Journal of Biological Engineering 3(19): doi:10.1186/1754-1611-3-19. Cooling, M. T., Rouilly, V., Misirli, G., Lawson, J., Yu, T., Hallinan, J. & Wipat, A. (2010). Standard virtual biological parts: a repository of modular modeling components for synthetic biology. Bioinformatics 26: 925 - 931. Knight, T. (2003). Idempotent vector design for standard assembly of Biobricks. In Knight, T. MIT Synthetic Biology Working Group Technical Report 0 http://hdl.handle.net/1721.1/21168. . Marchisio, M. A. & Stelling, A. (2008). Computational design of synthetic gene circuits with composable parts. Bioinformatics 24(17): 1903 - 1910. Richardson, L. (2007). RESTful Web Services. Pragma. Rodrigo, G., Carrera, J. & Jaramillo, A. (2007). Genetdes: Automatic design of transcriptional networks. Bioinformatics 23(14): 1857 - 1858.
 "
"