Team:HokkaidoU Japan/Projects/PCR
From 2010.igem.org
English / 日本語
PCR Based Assembly Protocol
Sometimes standard protocol fails to produce plasmid or BioBrick is toxic to host cell and can't by amplified. In moments like these PCR is handy. PCR is also great because it doesn't have many steps, the more steps the more ways to fail.
When amplifying BioBricks primers which anneal to prefix and suffix are used. Good thing about them is that they are universal so with only 2 you can amplify all BioBricks. But unlike miniprep product BioBrick amplified like this don't produce 2 distinct bands when digested. So you are left wondering if digestion went well.
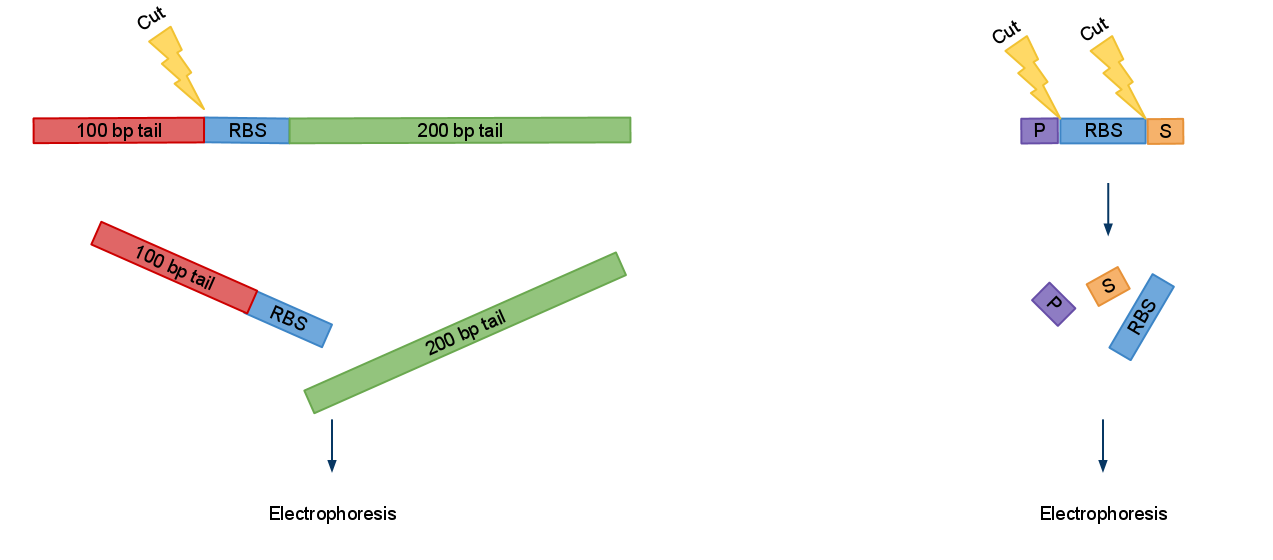
We created a set of primers which solves this problem. You can judge if digestion was successful by presence of approximatively 100 bp or/and 200 bp bands and significant change in PCRed fragments length.
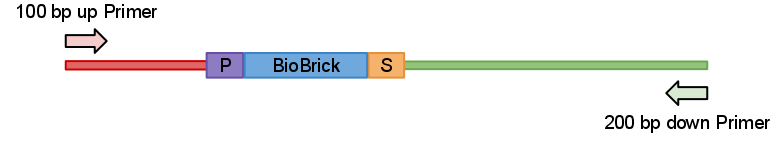
So idea behind this primers quite simple. We just designed primers which anneal 100 bp upstream and 200 bp down stream from the part like in Fig.1 bellow.
Now when digested PCRed fragment produces distinct approx 100 bp or/and 200 bp fragment depending on cut sites. Knowing this you can continue to the next step with the sure and light heart (see Fig.2). This is crucial when you take into account that many participants have limited at the bench experience before hand.
As everyone have guest by now these primers anneal to plasmid regions flanking the BioBrick. And there are more than 40 different primers. But not all of them are quite so different in fact all 7 registries assembly plasmids share the same sequence on the BioBrick flanking regions. And few others do also, bringing the number up to about 10.
And with the new registry requirement to submit all BioBricks in pSB1C3 plasmid present the possibility that in near future many if not all plasmids could be supported by this primers making assembly easier.
Key Components of PCR Based Assembly Protocol
- Reliable proofreading PCR enzymes like KOD Plus NEO
- It's 80 times more accurate than taq (At the moment we are not endorsed by producer : TOYOBO)
- Digestion Visualization Primers (DV primers)
- A program to calculate mixes for digestion and ligation
About the program to calculate mixes
We made simple program using excel to calculate restriction enzyme digestion and ligation mixes. One only needs to choose:
- 2 part to ligate
- Primers used in PCR
- DNA concentration after PCR
- Part and plasmid ratio
And amount for other reagents will be displayed automatically. This spares everyone from mundane calculations and help to conserve reagents. This program was made only for 2010 kit and currently isn’t available publically. But we feel the need for program like this to be on BioBrick registry page and could do calculations for all the parts in the registry. This is our proposal for improving efficiency of part assembly. Please contact us if you have any suggestions.
Some other cool thing that you can do with these primers
1)When you got small amount of plasmid DNA (like the Red DNA of the Kit) you can go from it to ligation the same day by PCR. And using these primers you can make sure that everything is going well. This good to know when you want to use a part you didn't plan for and you need it fast.
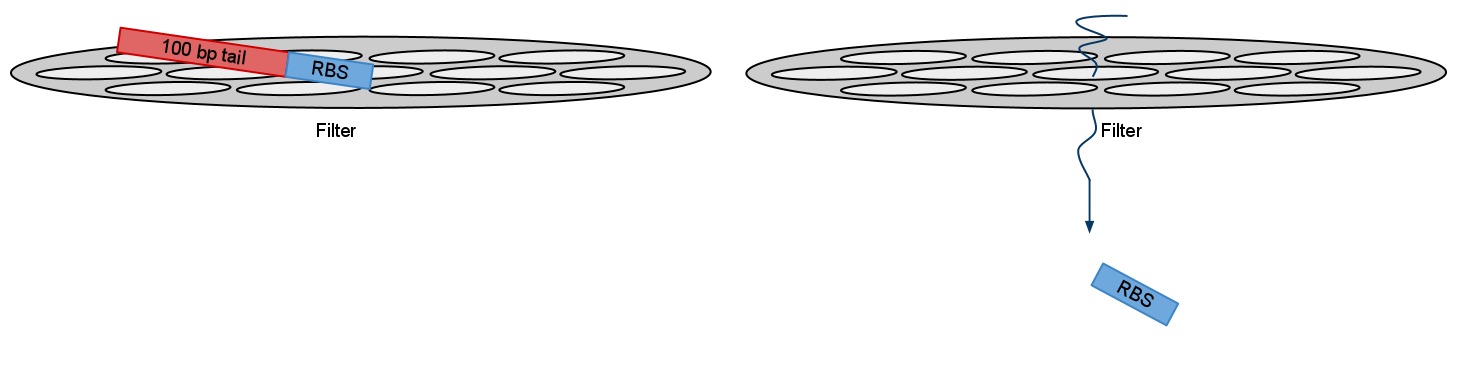
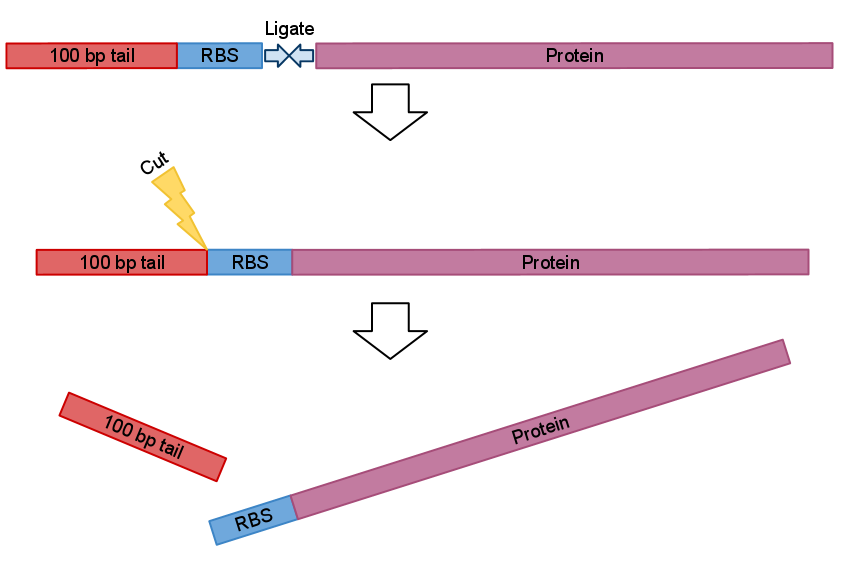
2)When using PCR parts size also comes into question. Small pieces like RBS or NLS signal are so small that they go right through purification filters. Purification kit suggested recovery rate for these parts is as small as 26% (for 50bp). Although we ordered oligos for RBS we used and NLS signals was included inside primers we suggest that in theory it is possible to use visualization primers to adding more bulk to PCRed part thus increasing it’s recovery rate.
After electrophoresis gel extraction needs to be performed. But recovery rate form small parts like RBS negligible. By adding bulk you increase recovery rate for filters. And bands should show in gel
After recovery and ligation with the part of your choice you just cut the bulk.
 "
"