Team:TU Delft/15 July 2010 content
From 2010.igem.org
Contents |
Lab work
Ordered DNA + Solvent Tolerance and Hydrocarbon Sensing
Some of transformations of yesterday containing the different ligations gave colonies (1 a 2 per plate). We picked as many colonies as possible of every plate and performed a colony PCR overnight to check which colonies contained the right insert. At the same time we grow them overnight in 5 mL LB medium containing the appropriate antibiotic.
Emulsifier
The results from the Colony PCR were not conclusive yesterday. That is why we decided to isolate the plasmid using Birnboim protocol. We used 3 mL of the bacterial cells to make -80 °C stocks.
The following plasmid concentrations were obtained:
| BioBrick | Composed of | Concentration (ng/μL) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398203 K398203] | OprG-B0015 (6) | |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398203 K398203] | OprG-B0015 (7) | |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398204 K398204] | AlnA-B0015 (11) |
The plasmids were subsequently digested with an enzyme that characteristic digest for all different plasmids. The plasmids were digested with HindIII. This should yield 3 bands in the lane with the plasmid pSB1T3 containing J04450 (neg control), 1 band in the lane with AlnA-B0015 and 2 bands in the lane with OprG-B0015 on gel.
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA |
| 1 | 1.5 μg pSB1T3 | 2 (BioLabs) | ✓ | ||
| 2 | 1 μg OprG-B0015 (6) | 2 (BioLabs) | ✓ | ||
| 3 | 1 μg OprG-B0015 (7) | 2 (BioLabs) | ✓ | ||
| 4 | 1 μg AlnA-B0015 (11) | 2 (BioLabs) | ✓ |
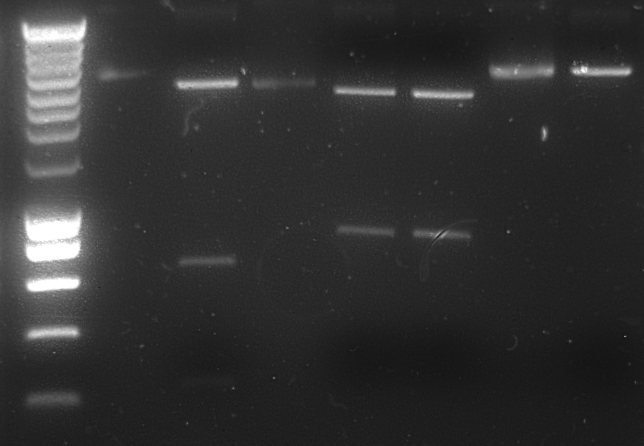
1% Agarose gel of digestion products:
Lane description:
| # | Description | Expected Length (bp) | Status | Remarks |
| M1 | SmarLadder marker | n/a | n/a | |
| 1 | Undigested pSB1T3 | 3507 | ✓ | |
| 2 | Digested pSB1T3 | 1972, 1176 | ✗ | |
| 3 | Undigested OprG-B0015 (6) | 3285 | ✓ | |
| 4 | Digested OprG-B0015 (6) | 1828, 1475, 222 | ✓ | |
| 5 | Digested OprG-B0015 (7) | 1828, 1475, 222 | ✓ | |
| 6 | Undigested AlnA-B0015 (11) | 3648 | ✓ | |
| 7 | Digested AlnA-B0015 (11) | 3511 | ✓ |
Characterization of Anderson RBS sequences
Fluorescence measurements attempt #2: 1. Last night's cultures of TOP10 carrying K398500, K398501, K398502, K398503, K398504 and I13401 were measured for biomass (OD600) and diluted to an OD600 of about 0.1 (see exact OD600 measurement of dilutions in table below).
| # | OD600 |
| K398500 | 0.102 |
| K398501 | 0.108 |
| K398502 | 0.108 |
| K398503 | 0.107 |
| K398504 | 0.098 |
| I13401 | 0.109 |
2. Aliquots of 175 μL of the dilutions were pipetted into the wells of a 96-wells plate in three-fold. The plate was covered with a transparent film to prevent evaporation.
3. The fluorescence and OD600 were measured for 16.30 hours with intervals 10 minutes by means of the Biotek Synergy plate reader with the excitation filter set at 485nm and the emission filter at 520nm.
Assembly of reference construct: K081005 yielded no transformants and was thus transformed a second time on ampicillin plates. Transformants of J23100 were single colony PCRed (see gel image below) and used as inoculate for 5 mL of LB for later plasmid isolation and -80 °C stocks.
The PCR amplification product of K081005 was digested using EcoRI and SpeI. 2 μg of I13401 in pSB1A2 was digested using EcoRI and XbaI.
Positive control Transformants of I13522 were single colony PCRed (see gel image below) and used as inoculate for 5 mL of LB for later plasmid isolation and -80 °C stocks.
1% Agarose gel of scPCR product:
Lane descriptions:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| M1 | SmartLadder marker | n/a | n/a | n/a | |
| 1 | transformant #1 of J23100 | G00100 + G00101 | ✔ | ||
| 2 | transformant #2 of J23100 | G00100 + G00101 | ✔ | ||
| 3 | transformant #1 of I13522 | G00100 + G00101 | ✔ | ||
| M2 | EZ Load Ladder (5 μL) | n/a | n/a | n/a |
From the gel it can be concluded that the transformants carry the desired plasmids with the proper BioBrink inserts.
 "
"