Team:TU Delft/Project/alkane-degradation/results/ALDH
From 2010.igem.org
Characterization of the ALdehyde DeHydrogenase system
Assay based on growth (Aldehydes as sole C-source)
The recombinant strains Escherichia coli 029A ([http://partsregistry.org/Part:BBa_K398029 BBa_K398029] on the plasmid pSB1A2) and Escherichia coli 030A ([http://partsregistry.org/Part:BBa_K398030 BBa_K398030] on the plasmid pSB1A2) were culture on M9 using Octanal and Dodecanal as sole carbon sources. They didn't show visible growth after 48 hours. No further experiments using this protocol were performed afterwards.
Resting cell assays
- We grew 'Escherichia coli 029A, Escherichia coli 030Aand Escherichia coli negative control (Biobrick BBa_J13002 on plasmid pSB1A2 ) in 50 mL of M9 medium with glucose and CAS aminoacids.
- The cells were harevested when the O.D. at 600nm was around 0.3; they were spun down at 4000 rpm, for 10 min at 4ºC. And the resting cell assays were prepared according to the standard protocol
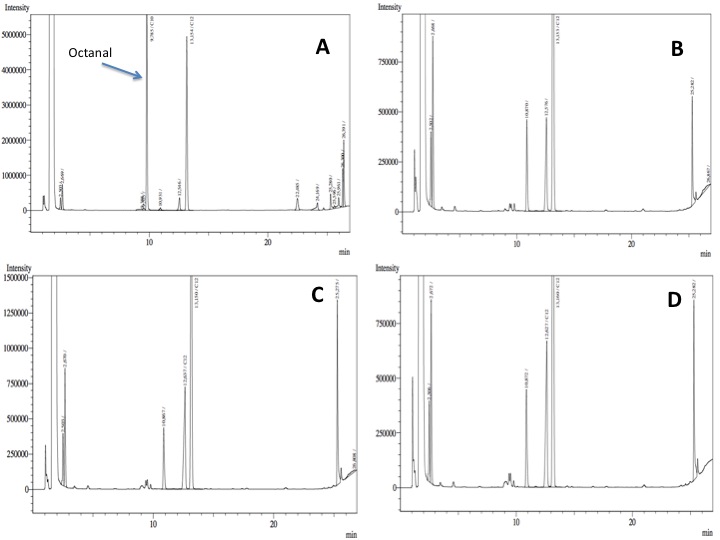
- After an overnight incubation at 37ºC with the substrate (octanal), the organic phase was extracted using 3 mL of ethyl acetate (dodecane was used as internal standard). We tried to determine production of alkanoic acids by Gas Chromatography measurements. The chromatograms are shown below:
- According to our results there was a peak reduction for octanal, however we were worried because of the fact that we didn't see the peak of the product (dodecanoic acid) appearing. It could be that the cells are just storing the product inside the cell. The experiment required to much time in order to get more results: the extractions, preparation of triplicates, cultures, cell suspensions... etc. it was a work that would require an effort of all the team for a couple of weeks. We decided to go for something easier and faster to measure; since we had some experience with the NADH determinations that was our choice.
Enzyme activity assay
Cells were cultured in 50mL of LB medium and harvested when the O.D. 600nm of the culture was between 0.5-0.8. Two different cultures of each recominant strain and the negative control were prepared.
Cytoplasmic proteins were extracted using our standard protocol. A standard curve for protein quantification was prepared using Bradford.
The total protein of each sample was quantified using 20uL of cell extract.
The aldehyde dehydrogenase activity was measured using the standard protocol and Dodecanal as substrate.
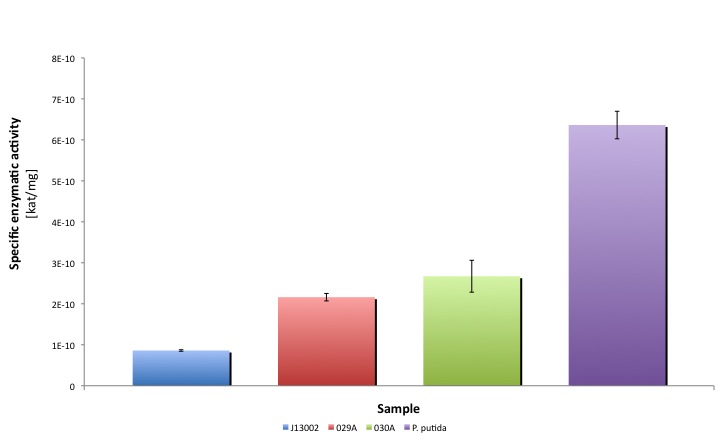
After the data treatment, the results that we obtained were the following:
| J13002 | 029A (1) | 029A (2) | 030A (1) | 030A (2) | P. putida | ||
|---|---|---|---|---|---|---|---|
| Specific activity | kat/mg | 8.604E-11 | 1.470E-10 | 2.853E-10 | 2.433E-10 | 2.912E-10 | 6.362E-10 |
| U/mg | 5.162E-03 | 8.820E-03 | 1.712E-02 | 1.460E-02 | 1.747E-02 | 3.817E-02 | |
| Standard deviation | 1.778E-12 | 6.187E-12 | 4.026E-11 | 3.539E-11 | 4.319E-11 | 3.361E-11 | |
| Relative to (J13002) | 100.00% | 170.86% | 331.59% | 282.79% | 338.46% | 739.41% | |
| Relative activity (P. putida) | 13.52% | 23.11% | 44.84% | 38.25% | 45.78% | 100.00% | |
| T-test vs J13002 | - | 0.9827 | 0.9599 | 0.9764 | 0.9976 | 0.9976 | |
| T-test vs P. putida | 0.9999 | 1.0000 | 0.9998 | 0.9974 | 1.0000 | - | |
| Average activity [kat/mg] | 8.604E-11 | 2.161E-10 | 2.673E-10 | 6.362E-10 |
You can download a file with our raw data, results anda summary here: File:TUDelft ALDH results.xls
Protein expression
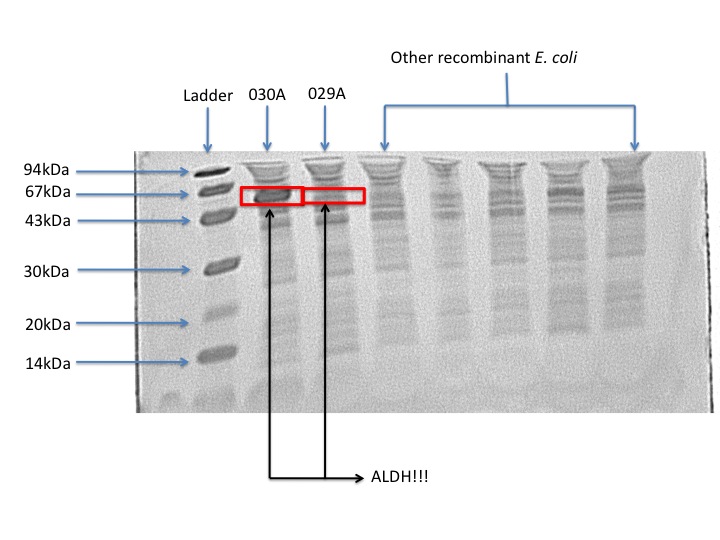
According to our protein gel, E. coli 030A overporduces ALDH. Whereas the protein production in E. coli 029A is barely visible. From this result we can conclude that overproduction is not necessary in order to obtain biological activity. From an overproduction of the enzyme the improvement is just 9% of the Pseudomonas putida activity. This could mean that there is protein biologically inactive, which is just a waste of cellular resources.
Conclusion
Our results suggest that the recombinant strains E. coli 029A and E. coli 030A functionally express our biobricks, we couldn't prove in vivo activity. However, it is clear from the statistical analysis performed that the expression of the biobrick [http://partsregistry.org/Part:BBa_K398006 BBa_K398006] under the promoter-rbs combination [http://partsregistry.org/Part:BBa_J13002 BBa_J23100]-[http://partsregistry.org/Part:BBa_J13002 BBa_J61117] increases the dodecanal dehydrogenase activity in E. coli cell extracts 2-fold; whereas the expression of the same protein using the part [http://partsregistry.org/Part:BBa_J13002 BBa_J13002] as promoter-rbs combo increases the same activity 3-fold. Moreover, the enzymatic activities measured for the constructs [http://partsregistry.org/Part:BBa_K398029 BBa_K398029] and [http://partsregistry.org/Part:BBa_K398030 BBa_K398030] were equivalent to 33.98% and 42.01% of the Pseudomonas putida aldehyde dehydrogenase activity, respectively.
In our resting cell experiment, apparently there is a decrease of the octanal peak, which may be due to the biological activity of our construct. However, it was not possible to see the formation of the expected product: octanoic acid. Other experiments like more resting cell assays (analyzing intracellular metabolites) or the use of C13 marked octanal may be useful in order to confirm and measure the in vivo biological activity of these parts.
- From the work published by Kato and co-workers in 2010 [http://www.springerlink.com/content/214116w7482469g0/fulltext.pdf], we knew that the purified enzyme has an activity equal to 0.63 U/mg, and according to the same study the tetradecanal dehydrogenase activity is 73 times higher than the octanal dehydrogenase; from the figure 5a of the cited paper we inferred that the dodecanal dehydrogenase activity is around 50% of the tetradecanal activity. Thus, the dodecanal dehydrogenase should be around 36.5 times the activity of octanal dehydrogenase which gives 22.995 U/mg pure ALDH.
- The same number expressed in kat/mg equals to 4e-7 kat/mg pure ALDH. Our cell extracts have dodecanal dehydrogenase activities of 2.15e-10 kat/mg protein cell extract (029A) and 2.67 e-10 kat/mg protein cell extract (030A). If we substract the normal E. coli activity, the net activities given by our biobricks are 1.301e-10 kat/mg protein cell extract (029A) and 1.812e-10kat/mg protein cell extract (030A). Which means that our strains had produced 3.253e-4 mg pure ALDH/mg protein cell extract (029A) and 4.53e-4 mg pure ALDH/mg protein cell extract (030A), respectively. Assuming a 70%(w/w) of protein content in E. coli, that means that a strain carrying pSB1A2 with the biobricks [http://partsregistry.org/Part:BBa_K398030 BBa_K398030] and [http://partsregistry.org/Part:BBa_K398029 BBa_K398029] will produce 0.0317% (w/w) and 0.0228% (w/w) of their total weight as ALDH protein.
- From the expression profile, we saw that the strain E. coli 029A produces less ALDH protein; however, the activity is comparable to the strain E. coli 030A which over produces ALDH, this means that the strain 029A spends more efficiently its cell resources and produces a highly active ALDH protein.
About our project and RBS characterization (link between projects)
- Originally, we decided to characterize the Anderson promoter family because we wanted to functionally express our proteins without stressing our cells. So far, with the biobrick [http://partsregistry.org/Part:BBa_K398029 BBa_K398029] we achieved an in vitro activity close to 34% of the activity reported for a natural alkane-degrading bateria. If someone else proves that our biobrick has the expected biological activity then that would mean that our design was successful.
- Nowadays, WE, as Synthetic Biologists, have the challenge of starting to produce parts that are being expressed in cells in the exact amount in order to confer biological activity. Over-expression (the standard protein expression method so far) is equal to waste; cells invest most of their resources for protein production (stressing the cells [http://www.springerlink.com/content/69k0pcbyc4l2qmfx/]); moreover, the production of inclusion bodies (aggregates of inactive protein) is reported when there is a high over-expression level [http://mbel.kaist.ac.kr/lab/research/protein_en1.html]. This means that high levels of protein over-expression will give as output large quantities of protein without biological function.
- With this project, we wanted to give other teams an insight about how much of specific proteins should be expressed in a cell in order to confer biological activity. Questions that arose at the beginning of our project were: How much of a protein should be expressed in order to have biological activity? Which promoter-rbs combo should be used in order to achieve a successful expression of a biologically active protein?. During the design phase (first two months), we didn't find answers to these questions and we decided to take the initiative and give the first steps towards finding answers to our questions.
- Some people may think that we didn't succeed on proving that our biobricks confer biological activity, however we feel that with this study we are giving the first steps on the way of engineering pathways using standard parts and rational expression of proteins. Some numbers about biologically active protein were given, in order to confirm our statements more studies are required and we suggest to use [http://partsregistry.org/Part:BBa_K398029 BBa_K398029] as a starting point, ADH ([http://partsregistry.org/Part:BBa_K398018 BBa_K398018]) is also a nice starting point because we confirmed that has biological activity; however its performance is really mediocre compared to a natural oil-degrading bacteria as Pseudomonas putida.
What would we measure if we had more time for this project?
In vivo activity tests: A comparison of in vitro activities maybe it is not interesting from the functional point of view, since those differences could be related to substrate affinity or discrepancies in the optimal pH. C13-labeled Dodecanal degradation measurements or resting cell assays or other kinds of studies are required in order to prove the in vivo activity of our proteins.
Fraction of Bt-ALDH expressed compared to the total amount of protein in the cell. We inferred the amount of ACTIVE Bt-ALDH in our cell extract, however this number is not accurate until we can measure the total amount of Bt-ALDH in the recombinant strain.
Expression of Bt-ALDH using other promoter-rbs combo. This will give us an insight about the optimum promoter-rbs combo required for the highest possible activity without causing cell stress.
If someone else reports that our numbers and statements are correct, then they will confirm by the very first time in the iGEM competition the amount of protein required for successful expression of an in vivo biologically active enzyme. We will look forward for anyone willing to do this job on 2011 ;-)

 "
"