Team:Stockholm/Modelling
From 2010.igem.org
| Line 3: | Line 3: | ||
---- | ---- | ||
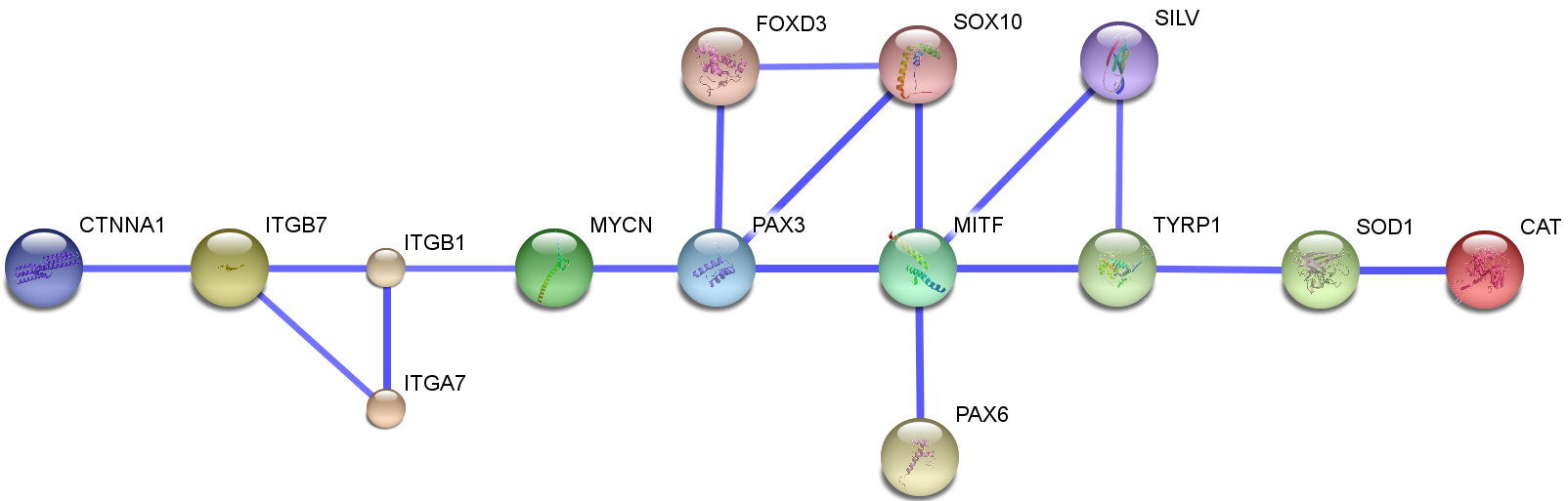
| - | [[Image:Net_Image_confidence_View_igem_stockholm_modelling_report.png|600px|thumb|center|Figure 1]] | + | [[Image:Net_Image_confidence_View_igem_stockholm_modelling_report.png|600px|thumb|center|Figure 1 Produced by STRING database Jensen et al. Nucleic Acids Res. 2009, 37(Database issue):D412-6]] |
Wet-lab members are working on several proteins. The idea behind why we chose these proteins was somehow vague, because of low amount of information we had on our wiki page. In this section we try to explain the reasons and ideas behind choosing those specific proteins. | Wet-lab members are working on several proteins. The idea behind why we chose these proteins was somehow vague, because of low amount of information we had on our wiki page. In this section we try to explain the reasons and ideas behind choosing those specific proteins. | ||
| - | As a starting point, we began with the article [http://www.ncbi.nlm.nih.gov/pubmed/18426409 Strömberg S et al. (2008)]. In this paper they identified some 859 genes as differentially regulated genes in melanocytes. These genes can be classified in several groups based on their possible role. These groups are: | + | As a starting point, we began with the article [http://www.ncbi.nlm.nih.gov/pubmed/18426409 Strömberg S et al. (2008)]. In this paper they identified some 859 genes as differentially regulated genes in melanocytes. These genes can be classified in several groups based on their possible role (Strömberg et al. 2008). These groups are: |
# Developing melanocytes | # Developing melanocytes | ||
# Melanin synthesis and delivering it using melanosomes | # Melanin synthesis and delivering it using melanosomes | ||
| Line 13: | Line 13: | ||
# Antigen presenting | # Antigen presenting | ||
| - | out of those genes, it seemed that [http://www.ncbi.nlm.nih.gov/gene/4286 MITF] is one of the most | + | out of those genes, it seemed that [http://www.ncbi.nlm.nih.gov/gene/4286 MITF] is one of the most regulated and regulating genes in vitiligo disease. This was the first block of the map. . The MITF promoter is targeted by several transcription factors that are important in neural-crest development and signaling. Transcription factors implicated in the regulation of the MITF promoter include PAX3, cAMP-responsive element binding protein (CREB), SOX10, LEF1 (also known as TCF), one cut domain 2 (ONECUT-2) and MITF itself (Levy et al., 2006) and it also regulates both the survival and differentiation of melanocytes, and enzymes which are necessary for melanin production. (Levy et al., 2006) |
| + | |||
| + | [[Image:SU_igem_mitf_regulatory_site.jpg|200px|thumb|center|Figure 2 Regulators and transcription-factor binding sites on the MITF promoter. Levy et al., 2006] | ||
| + | |||
| + | |||
| + | |||
| + | to be continued... | ||
| - | |||
Revision as of 12:59, 31 August 2010
Wet-lab members are working on several proteins. The idea behind why we chose these proteins was somehow vague, because of low amount of information we had on our wiki page. In this section we try to explain the reasons and ideas behind choosing those specific proteins.
As a starting point, we began with the article [http://www.ncbi.nlm.nih.gov/pubmed/18426409 Strömberg S et al. (2008)]. In this paper they identified some 859 genes as differentially regulated genes in melanocytes. These genes can be classified in several groups based on their possible role (Strömberg et al. 2008). These groups are:
- Developing melanocytes
- Melanin synthesis and delivering it using melanosomes
- Cell adhesion
- Antigen presenting
out of those genes, it seemed that [http://www.ncbi.nlm.nih.gov/gene/4286 MITF] is one of the most regulated and regulating genes in vitiligo disease. This was the first block of the map. . The MITF promoter is targeted by several transcription factors that are important in neural-crest development and signaling. Transcription factors implicated in the regulation of the MITF promoter include PAX3, cAMP-responsive element binding protein (CREB), SOX10, LEF1 (also known as TCF), one cut domain 2 (ONECUT-2) and MITF itself (Levy et al., 2006) and it also regulates both the survival and differentiation of melanocytes, and enzymes which are necessary for melanin production. (Levy et al., 2006)
[[Image:SU_igem_mitf_regulatory_site.jpg|200px|thumb|center|Figure 2 Regulators and transcription-factor binding sites on the MITF promoter. Levy et al., 2006]
to be continued...
We start by looking at several databases how the PPI map should look like for the proteins of interest, from there we can make statements about predictions or make prove of concept. One of the main articles which looked into transcriptional profile in melanocytes from vitiligo patients is Strömberg S et al. Pigment Cell Melanoma Res. (2008). For the first phase of interaction map, focuse will be on MITF. For this case, I chose several genes which are differentially regulated from Strömberg S et al. Pigment Cell Melanoma Res. (2008). foxd3, pax3 and tyrp1 are chosen (Notes from August 6 and August 5).
 "
"