Team:Newcastle/10 August 2010
From 2010.igem.org
Shethharsh08 (Talk | contribs) (→Results from Nanodrop:) |
Shethharsh08 (Talk | contribs) (→Materials and Protocols) |
||
| Line 90: | Line 90: | ||
|Restriction digest EcoR1 and Pst1 | |Restriction digest EcoR1 and Pst1 | ||
|- | |- | ||
| - | !DNA | + | !DNA (in µl) |
| - | !EcoR1 | + | !EcoR1 (in µl) |
| - | !Pst1 | + | !Pst1 (in µl) |
| - | !10 times buffer | + | !10 times buffer (in µl) |
| - | !Water | + | !Water (in µl) |
|- | |- | ||
!5 | !5 | ||
Revision as of 12:56, 11 August 2010

| |||||||||||||
| |||||||||||||
Contents |
Gel extraction of rocF BioBrick components
Gel Electrophoresis for Amplified Pspac_oid promoter, Double terminator and Plasmid Vector pSB1C3
Aim
The aim of this experiment is to perform gel electrophoresis for the amplified fragments of Pspac oid promoter, double terminator and plasmid vector pSB1C3 and then perform gel extraction of the required bands.
Materials and Protocol
Please refer to: Gel electrophoresis.
Result
The experiment is divided in two separate gels. For plasmid vector pSB1C3, we used 1% agarose gel but for Psapc_oid and double terminator fragment we used 1.5% agarose gel for a better resolution. We used a 1Kb DNA ladder (Promega) for the gel containing plasmid vector while we used a 100bp DNA ladder (Promega) for the gel containing Pspac_oid and double terminator fragment.
| Pspac_oid pormoter | Double Terminator | Plasmid Vector pSB1C3 | |
|---|---|---|---|
| Size of the Fragment (in bp) | 148 approx. | 116 approx. | 2072 approx. |
Table 1: Table represents the sizes of the Pspac_oid fragment, Double terminator and plasmid vector pSB1C3 represented as bands on the gel in their respective lanes.
Discussion
We found bands of appropriate sizes in their respective lanes.
Conclusion
Gel electrophoresis was successful and now we would be doing gel extraction of all the six fragments (including the three fragments on which we did gel electrophoresis today) fragments.
Gel extraction of all 6 fragments for rocF
Aim
The aim of this experiment is to perform gel extraction of the bands containing the amplified fragments of the Pspac oid promoter, double terminator and plasmid vector pSB1C3.
Materials and Protocol
Please refer to: Gel extraction.
Result
The experiment is divided in two separate gels. For plasmid vector pSB1C3, we used 1% agarose gel but for Psapc_oid and double terminator fragment we used 1.5% agarose gel for a better resolution. We used 1Kb DNA ladder (Promega) for the gel containing plasmid vector while we used 100bp DNA ladder (Promega) for the gel containing Pspac_oid and double terminator fragment.
| Pspac_oid pormoter | Double Terminator | Plasmid Vector pSB1C3 | |
|---|---|---|---|
| Size of the Fragment (in bp) | 148 approx. | 116 approx. | 2072 approx. |
Table 2: Table represents the sizes of the Pspac_oid fragment, Double terminator and plasmid vector pSB1C3 represented as bands on the gel in their respective lanes.
Discussion
We found bands of appropriate sizes in their respective lanes.
Conclusion
Gel electrophoresis was successful and now we would be doing gel extraction of all the six fragments (including the three fragments on which we did gel electrophoresis today) fragments.
Mixing the Gibson reaction buffers
Aim
The aim of this experiment is to mix the 5X isothermal reaction buffer and 1.33X Master Mix required for Gibson DNA assembly. Please see the Gibson cloning protocol.
lacI and pVeg
Aims
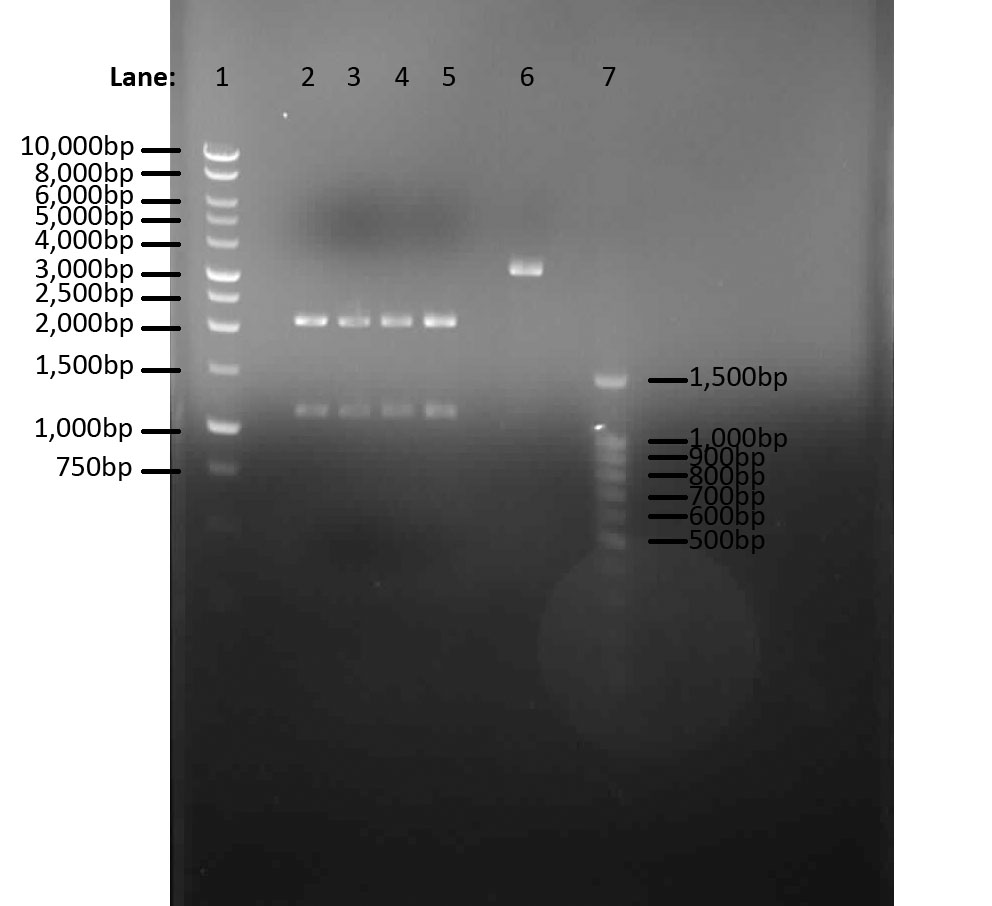
We aim to extract pVeg and lacI from transformed E.coli DH5α which was cultured in LB broth overnight(see 09.08.2010)check the concentration of DNA using nanodrop and perform a restriction digest so we can run the samples on a gel to check the insert sizes.
Materials and Protocols
| Restriction digest EcoR1 and Pst1 | ||||
| DNA (in µl) | EcoR1 (in µl) | Pst1 (in µl) | 10 times buffer (in µl) | Water (in µl) |
|---|---|---|---|---|
| 5 | 0.5 | 0.5 | 1 | 3 |
See protocols for materials
Results
Results from Nanodrop:
| lacI 1 | lacI 2 | lacI 3 | lacI 4 | pVeg | |
|---|---|---|---|---|---|
| Concentration of DNA ng/µl | 122.5 | 139.4 | 130.0 | 150.0 | 155.0 |
Table 4: Table represents the concentrations of different amplified fragments in ng/µl.
Results from PCR:
| Lane | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1,000bp marker | lacI 1 | lacI 2 | lacI 3 | lacI 4 | pVeg | 100bp marker |
Subtilin Immunity BioBrick
Aims
Lab work today will involve the rehydration of the primers that have arrived today. The six primers ordered are Primer 1 P-1, Primer 2 P-2, Primer 1 S-1, Primer 2 S-1, Primer 1 T-1 and Primer 2 T-1. The other primers are Primer 1 V-1 and Primer 2 V-1, which we already have in stock, therefore no rehydration will be needed. (Primer 1 = Forward primer and Primer 2 = Reverse primer).
After the primers are ready, Phusion PCR will be performed, using four different template DNA: Plasmid Vector, Promotor & RBS, spaIFEG gene cluster and Double Terminator.
For more information about the subtilin immunity BioBrick, please see the cloning strategy for subtilin immunity (Link)
Materials and Protocol
Please refer to the DNA Re-hydration protocol.
Please refer to the Phusion PCR protocol.
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Plasmid Vector | PSB1C3 | P1V1 forward | P2V1 reverse | 53.3 | 2046 approx. or +? | 70 |
| 2 | Promoter and RBS (pVeg-SpoVG) | BioBrick Bba_K143053 | P1P1 forward | P2P2 reverse | 51.7 | 139 approx. or +? | 15 |
| 3 | spaIFEG Gene Cluster | B. subtilis ATCC 6633 | P1S1 forward | P2S1 reverse | 46.0 | 2753 approx. or +? | 110 |
| 4 | Double terminator | pSB1AK3 consisting BBa_B0014 | P1T1 forward | P2T1 reverse | 50.9 | 116 approx. or +? | 15 |
Table 5: This table shows the four different Phusion PCR reactions that were carried out today. If this is successful, the four parts can be ligated together for the construction of subtilin immunity BioBrick with the help of Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Thus the extension time of each and every PCR reaction is slightly different.
- Please note: The newly arrived primer tubes have to be handled with extra caution, because they will be the main stocks which working stock solutions will be made from. Therefore, gloves have to be worn, as well as preventing any contamination. Water will be used in order to liquefy the primers and the water used will be from Pure Lab Distilled Water.
Results, Discussion and Conclusion
For results, discussion and conclusion, please refer to the Lab book on 11 August.
Go back to our main Lab book page
 
|
 "
"