Team:SDU-Denmark/labnotes
From 2010.igem.org
(Difference between revisions)
(→Lab notes (7/12 - 7/18)) |
(→Lab notes (7/12 - 7/18)) |
||
| Line 12: | Line 12: | ||
==== Exp. 1 ==== | ==== Exp. 1 ==== | ||
<br> | <br> | ||
| - | ''Methods:'' [https://2010.igem.org/Team:SDU-Denmark/protocols# | + | ''Methods:'' Miniprep, NanoDrop and gel electrophoresis <br><br> |
| + | ''protocols:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#MP1.1 MP1.1] <br><br> | ||
''Notes:'' We used 9 ml ON culture (Lag phase cells), loaded 2ul sample and 8ul agarose loading dye on a 1,5% gel,used a DNA ladder mix (100-10000 nucleotides) as marker <br><br> | ''Notes:'' We used 9 ml ON culture (Lag phase cells), loaded 2ul sample and 8ul agarose loading dye on a 1,5% gel,used a DNA ladder mix (100-10000 nucleotides) as marker <br><br> | ||
''Results:'' Nanodrop: Tube 1: 30.7 mg/ul Tube 2: 27.4 mg/ul <br><br> | ''Results:'' Nanodrop: Tube 1: 30.7 mg/ul Tube 2: 27.4 mg/ul <br><br> | ||
| Line 28: | Line 29: | ||
==== Exp. 3 ==== | ==== Exp. 3 ==== | ||
| - | ''Methods:'' | + | ''Methods:'' MiniPrep, NanoDrop and gel electrophoresis. <br><br> |
| + | ''protocols:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#MP1.1 MP1.1] <br><br> | ||
''Notes:'' We used 10 ml af a culture in Log phase (1ml cells from an ON culture + 9ml LB medium, incubated at 37 degrees celcius for 4 hours), loaded 4ul sample and 4ul agarose loading dye on a 1,5% gel,used a DNA ladder mix (100-10000 nucleotides) as marker. <br><br> | ''Notes:'' We used 10 ml af a culture in Log phase (1ml cells from an ON culture + 9ml LB medium, incubated at 37 degrees celcius for 4 hours), loaded 4ul sample and 4ul agarose loading dye on a 1,5% gel,used a DNA ladder mix (100-10000 nucleotides) as marker. <br><br> | ||
''Results:'' Nanodrop: Tube 1: 38.7 mg/ul Tube 2: 32.4 mg/ul. <br><br> | ''Results:'' Nanodrop: Tube 1: 38.7 mg/ul Tube 2: 32.4 mg/ul. <br><br> | ||
| Line 37: | Line 39: | ||
=== Polyferation of FlhDC, FlhD and FlhC genes === | === Polyferation of FlhDC, FlhD and FlhC genes === | ||
<br> | <br> | ||
| - | ''Methods:'' | + | ''Methods:'' PCR and Gel electrophoresis.<br><br> |
| + | ''Protocols:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.2 CHP1.2] <br><br> | ||
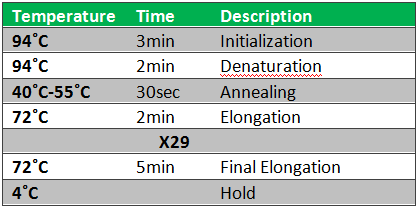
''Notes:'' We decided to test our primers on previously purified cromosomal DNA. Examination of the primers showed that the FlhC reverse primer had a melting temperature of only 45˚C. Therefore we decided to run the samples on a gradient PCR. Simultaneously, we prepared 2 extra samples, isolating FlhD and FlhC, respectively. We did this because we wanted to see if our problems were caused because the combined gene-sequence was to long (932bp).<br> | ''Notes:'' We decided to test our primers on previously purified cromosomal DNA. Examination of the primers showed that the FlhC reverse primer had a melting temperature of only 45˚C. Therefore we decided to run the samples on a gradient PCR. Simultaneously, we prepared 2 extra samples, isolating FlhD and FlhC, respectively. We did this because we wanted to see if our problems were caused because the combined gene-sequence was to long (932bp).<br> | ||
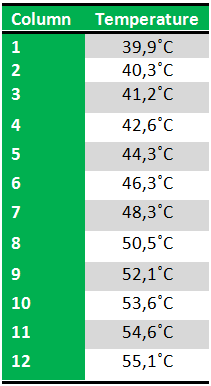
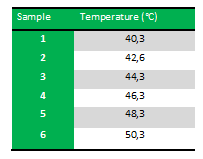
Because we just wanted to test our primers in this PCR, we used Taq polymerase, because although it doesn’t proofread, it is remarkably cheaper than Pfu. On the [http://www.fermentas.com/en/products/all/pcr-qpcr-rt-pcr/standard-pcr/ep028-taq-dna-native Fermentas homepage] we found that the annealing temperature for Taq is Tm-5 , which in this case means 40˚C. However, Taq polymerase is not very effective at temperatures under 50˚C so we designed the gradient to lies between 40 and 55˚C. The PCR machine chose the following temperatures: | Because we just wanted to test our primers in this PCR, we used Taq polymerase, because although it doesn’t proofread, it is remarkably cheaper than Pfu. On the [http://www.fermentas.com/en/products/all/pcr-qpcr-rt-pcr/standard-pcr/ep028-taq-dna-native Fermentas homepage] we found that the annealing temperature for Taq is Tm-5 , which in this case means 40˚C. However, Taq polymerase is not very effective at temperatures under 50˚C so we designed the gradient to lies between 40 and 55˚C. The PCR machine chose the following temperatures: | ||
| Line 58: | Line 61: | ||
<br> | <br> | ||
''Date'': july 12th <br><br> | ''Date'': july 12th <br><br> | ||
| - | ''methods'': [https://2010.igem.org/Team:SDU-Denmark/protocols# | + | ''methods'': PCR and gel electophoresis <br><br> |
| + | ''Protocols:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.2 CHP1.2] <br><br> | ||
''Notes'': The purified chromosomal DNA was tested with Pfu; a polymerase with proofreading ability. The DNA was from the E. coil strain MG 1655 (tube 17) The annealing temperature was used fore the PCR was 44˚C. We decide use this annealing temperature while it is in the middle of the temperature span we successful used in the last experiment. Further we increas the elongation time to 1 min and 30 sec. Other than these changes we followed the PRC program in the protocol. the PCR product was run on a 1,5% agarose gel. <br><br> | ''Notes'': The purified chromosomal DNA was tested with Pfu; a polymerase with proofreading ability. The DNA was from the E. coil strain MG 1655 (tube 17) The annealing temperature was used fore the PCR was 44˚C. We decide use this annealing temperature while it is in the middle of the temperature span we successful used in the last experiment. Further we increas the elongation time to 1 min and 30 sec. Other than these changes we followed the PRC program in the protocol. the PCR product was run on a 1,5% agarose gel. <br><br> | ||
''results'': Unfortunately the experiment did not give any results on the gel. <br><br> | ''results'': Unfortunately the experiment did not give any results on the gel. <br><br> | ||
Revision as of 05:33, 14 July 2010
 "
"