Team:ETHZ Basel/Biology
From 2010.igem.org
(→generation of BioBricks) |
|||
| Line 1: | Line 1: | ||
{{ETHZ_Basel10}} | {{ETHZ_Basel10}} | ||
| + | <html><div id="body-box"></html> | ||
== Overview == | == Overview == | ||
| Line 48: | Line 49: | ||
== Bringing ''E. lemming'' to life == | == Bringing ''E. lemming'' to life == | ||
| + | |||
| + | <html></div></html> | ||
Revision as of 20:57, 6 September 2010
Overview
The goal of the iGEM project 2010 of ETH Basel "E.lemming" is to control the tumbling frequency of E. coli. This is achieved by spatially localizing certain elements of the chemotactic network (Che proteins) and thus affecting the activity of their downstream partners.
Reversible localization is achieved by the light-inducible PhyB-PIF3 system detected in plants. PIF3 will be fused to a Che protein and PhyB to a localized anchor within the cell. Red light illumination converts PhyB Pr to Pfr facilitating PhyB-Pif3 interaction and therefore spatially separating the Che protein from its binding partner. Upon far red light stimulus, Pfr converts back to Pr resulting in the dissociation of PhyB-Pif3 and free diffusion of the Che protein.
As modeling of the chemotactic network did not give a clear answer which Che protein should be attacked, several combinations will be investigated.
In view of the chemotactic proteins this includes CheY, CheB and CheR and concerning the localizer MreB (localization to the membrane), tetR (localization to the plasmid via tetO) and trigger factor (localization to the ribosome).
The goal of the wet lab team is to implement this localization system into E. coli.
Cloning Strategy
As we plan to generate several fusion proteins with different linkers, we decided to use the cloning strategy BBF RFC 28: A method for combinatorial multi-part assembly based on the Type IIs restriction enzyme AarI (http://dspace.mit.edu/handle/1721.1/46721). The advantage of this strategy is that we can clone up to 3 different inserts into one vector simultaneously in a 96 well format.
generation of BioBricks
All utilized parts will be generated by PCR and subcloned into the storage vector pSEVA132 (Victor de Lorenzo's lab, KanR, BBR1 ori) allowing blue white screening. The working process for the generation of the subparts is as follows:
1. Ordering of primers (if template is available)
2. PCR
3. clean-up of PCR product
4. Ligation into storage vector
5. Transformation
6. Blue-white screening
7. Sequencing
Due to the presence of rare codons in the sequence of PhyB and Pif3, these two genes will be ordered from GenArt. However, as synthesizing takes several weeks, expression of the wild-type gene of these two proteins will be tested and if satisfying proceeded with these constructs.
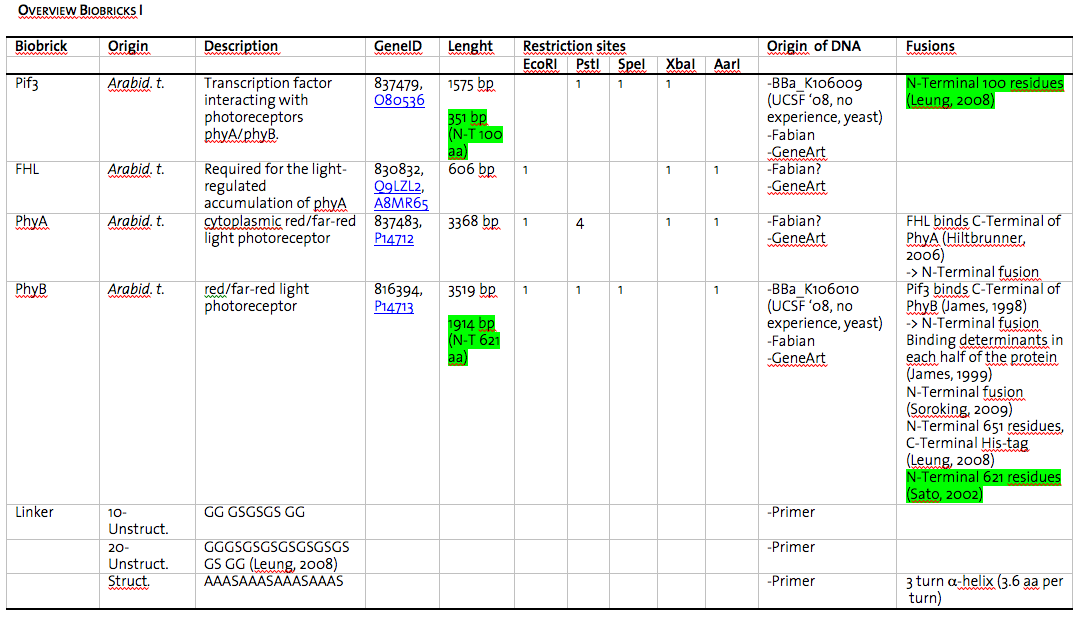
In this picture all the details (origin of DNA, length of gene, restriction sites, sources) about the BioBricks generated are displayed.
generation of fusion proteins
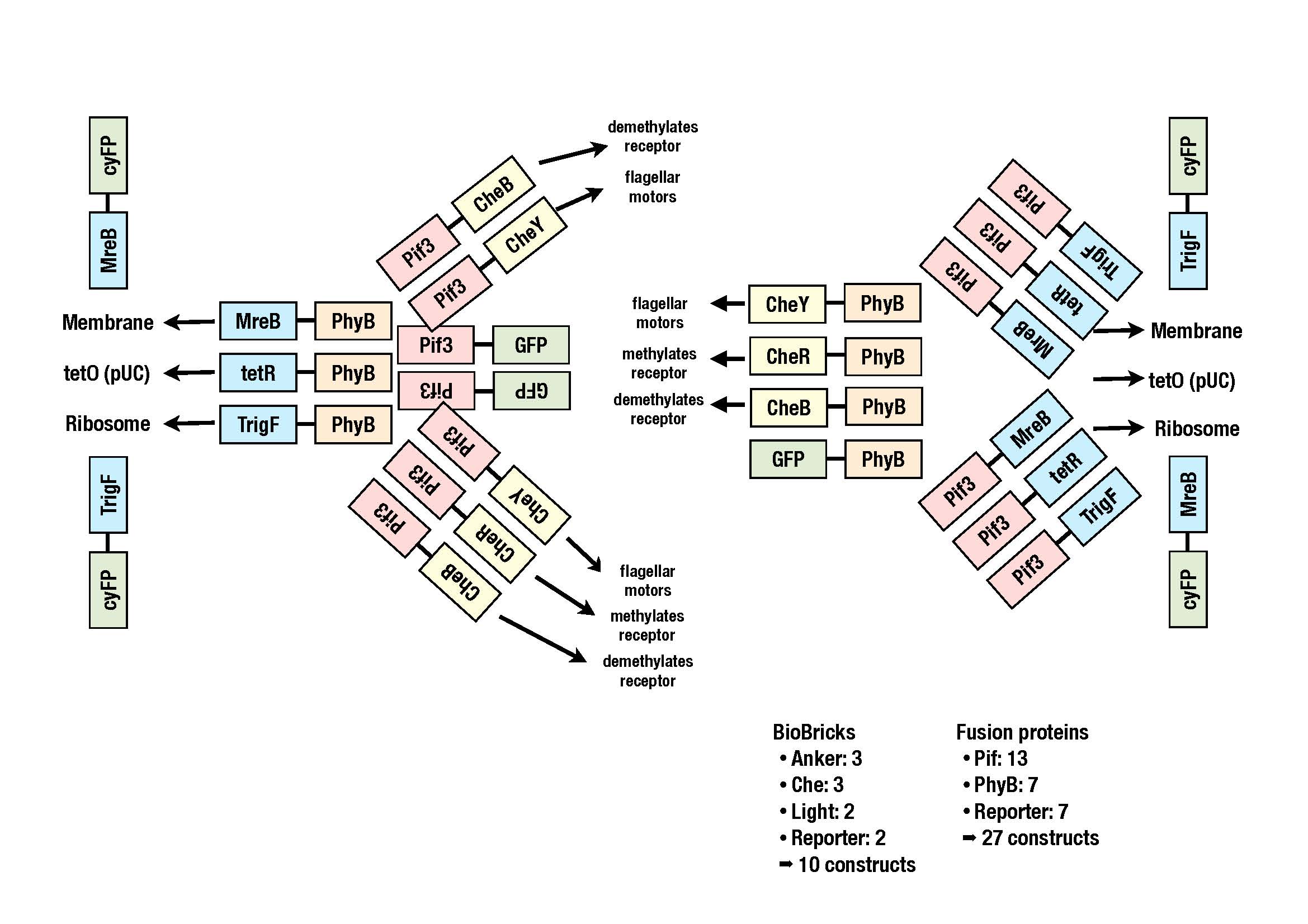
The image shows all the constructs we plan to clone.Once the BioBricks in the storage vectors are available, the fusion proteins in the acceoptor vector will be generated according to the cloning strategy BBF RFC 28.
Functionality assays
The constructs will have to be tested for the following properties:
- Che protein fusion: Influencing of chemotactic network
- Localizer fusion: Spatial localization
- PhyB-Pif system: Activation by light
Chemotactic Functionality
Is the Che fusion protein is still functional?
Possible assays to investigate the functionality:
Localization
Is the localizer spatially separating PIF3 or PhyB to a certain area within the cell?
Fluorescence microscopy will be utilized to answer this question. Therefore, fusions of the anchor to fluorescent proteins (cyfp, gfp) have to be generated.
 "
"